Curation in context: A glimpse into COSMIC v99
29 Nov 2023
At its most basic level, all cancer research is motivated by the potential to improve patient outcomes, no matter how far removed from the bedside the research may seem. This unifying goal has been responsible for countless medical and technological advancements dating back hundreds of years. From early attempts at surgical tumour removal, to the rise of radiation therapy in the late nineteenth century, progress was slow at first, but after Watson and Crick’s discovery of DNA structure in 1952, a whole new branch of research emerged. Genetic research went from strength to strength in the late twentieth century. It took only 20 years from the discovery of the first oncogene in 1970, for the human genome project to be launched. Over a decade later the first sequence of a human genome was produced in 2003, and today, Ilumina offers sequencers that can generate more than 20,000 whole genomes a year.
This boom in technology, development and understanding has resulted in an extremely data driven era of cancer research. Without curation into accessible repositories, the masses of potentially crucial data produced as a result of this ‘boom’ can find itself lost in the vast sea of literature. With this in mind, for v99, the COSMIC team has dedicated itself to the expert curation of a range of integral COSMIC data. This focus has included 7 expertly curated genes, 6 census genes, 8 cancer hallmark genes, plus a new resistance gene drug pair.
Expertly curating genes
So, what makes a gene ‘expertly curated’? The COSMIC curation team is made up of postdoctoral level scientist curators who are dedicated to manually interpreting data from peer reviewed publications. This manual approach allows for an extremely high level of quality control, allowing curators to pick out error inconsistencies in publications that may go unnoticed through a systematic approach.
The search for this information begins where countless university assignments, theses and groundbreaking research has before: a broad search of relevant data on PubMed. More specifically, the team will often begin looking for mutation data from a specific gene (an example search is: (ras OR genes, ras) AND human AND mutation). The genes selected to be investigated are typically ones for which there are no existing databases, but are included in an assembled list of genes that are somatically mutated and causally implicated in human cancer.
Papers identified as potentially containing data of interest are then examined in full before up to 45 different data points per sample are pulled out, including: Tumour/ tissue type, sample, mutation and individual information. Any papers containing data that does not meet our quality standards will not be curated, but added to a list of additional references.
An example of the result of this meticulous work is the newly expertly curated gene for COSMIC v99, BAX: BCL2 Associated X, apoptosis regulator.
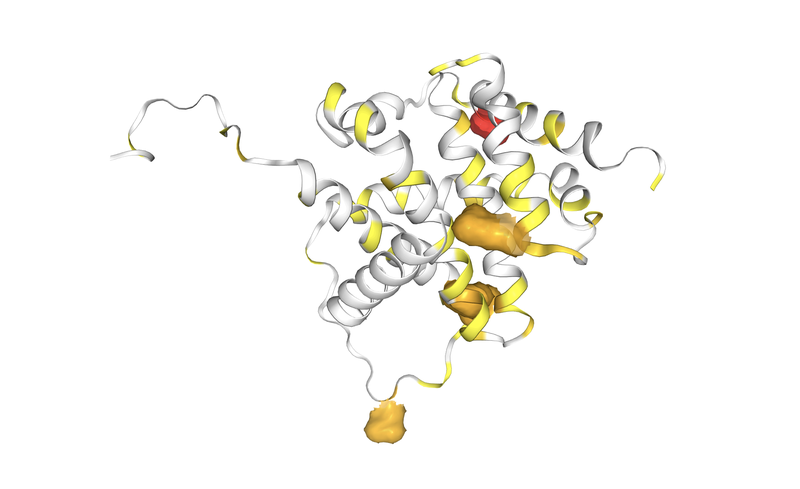
Image above: 3D protein model of BCL2 Associated X, apoptosis regulator protein via COSMIC 3D.
A short snippet of the information you could discover: Mutations in BAX are associated with many cancers with a particular prevalence in colorectal cancer, endometrial cancer and haematopoietic and lymphoid neoplasms. Many mutations occur within a poly (G) 8 tract within exon 3 and are associated with microsatellite instability, with around 90% of the new mutations curated for BAX being insertions or deletions in this region. The majority of these mutations are involved in cancers of the stomach and intestines. However, missense mutations in other parts of the BAX gene have also been curated in a broader spectrum of cancers including the haematopoietic or lymphoid cancers, cancers of the skin (especially malignant melanomas), liver and breast cancers.
Pieces of a puzzle
With over 6,800 distinct forms of human cancer recorded in COSMIC alone, it is important to remember that while deep analysis is needed, there is an expansive and diverse breadth of knowledge that needs to be addressed in equal measure. Like pieces of a puzzle, each data point has a role to play. Expert curated genes, mutational signatures, hallmarks annotations, these focus on mechanisms, causes and distribution of cancers, but how do we tackle these diseases? Our Actionability dataset keeps vigilant watch over efforts to combat cancer by curating current state of precision oncology, tracking treatment availability and trials in incredible detail.
Of course, treatment doesn’t always run smoothly. It is unfortunately quite common for a tumour to respond well initially, but for resistance to occur as time goes by. This leaves the curation of new resistance gene drug pairs, such as IDH1-Ivosidenib for COSMIC v99, crucial for patient outcomes. Ivosidenib is a drug often used, in part, to treat Acute Myeloid Leukaemia (AML). With thousands of patients diagnosed, and losing their life to AML yearly, there is an immense pressure to address any hindrance to treatment. FDA approved treatments, alternative treatment development, reasons for trial termination and much more, provide a perfect jumping off point for drug and treatment development. Churchill famously once said “Those that fail to learn from history are doomed to repeat it.’. A comprehensive understanding of past successes and failures is indispensable in any industry to optimise the work being done. Millions of patients are diagnosed with, and die of cancer every year, with the stakes this high, the optimisation of research is of the utmost importance in the battle against these diseases.
A data-driven future
When producing and analysing data, it can often be difficult to picture the consequences at a human level. This is where databases like COSMIC come in. Adaptation of curated data into analytical tools and accessible formats, allows researchers to gain actionable insights and offers real world context. Despite being crucial infrastructure in the data-driven race against countless diseases, databases are often launched, deprived of funding and then stagnate or disappear entirely. It is this that drives COSMIC’s dedication not only to curating gold-standard data, but to longevity. By focusing on integral data such as Cancer Gene Census updates and individual gene focuses, COSMIC v99 perfectly emulates this commitment to being a sustainable and reliable source of genomic data. It is only through a meticulously maintained balance of curation of new brand new information, and the revisiting of historical data that we can uphold the high standards we have been known and trusted for, for almost 20 years.