COSMIC Release v83
7 Nov 2017
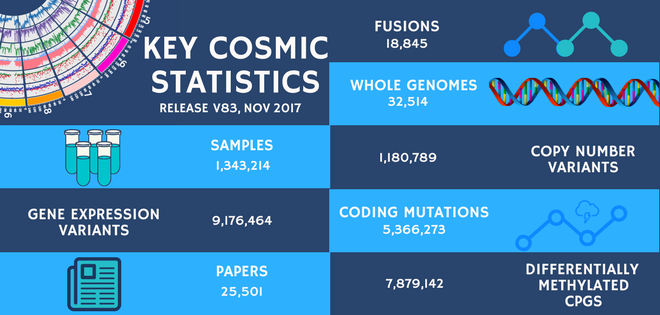
The November release (v83) is now live! New in this release we have 3 fully curated genes, a significant update to a fourth and a new fusion gene pair, as well as the incorporation of over 1000 new genomes from 13 systemic screen papers. The updates from the ICGC release 25 have also been included.
The new genes include: TGFBR2, transforming growth factor beta receptor 2, encodes a transmembrane member of the Ser/Thr protein kinase family which forms a heterodimeric complex with TGF-beta receptor type-1 and binds TGF-beta. TGFBR2 acts as a tumour suppressor gene in colorectal cancer, where its mutational inactivation is the most common genetic event affecting the TGF-beta signalling pathway, occurring in approximately 30% of these cancers. Genome studies have also identified TGFBR2 as a significantly mutated gene in cervical cancer, and head and neck squamous cell carcinoma. ERBB4, a comparatively less well understood member of the EGFR family of receptor tyrosine kinases, somatic mutations in ERBB4 are seen across various cancers including breast, lung and melanoma, and in various different regions of the gene. It has been proposed to act as both an oncogene and a tumour suppressor and is being investigated as a potential drug target. BCL9L, is a co-activator of Wnt/beta-catenin signalling. Here somatic loss-of-function alterations in BCL9L are frequent in aneuploid colorectal carcinoma but are also found in other tumour types at lower frequency. BCL9L has been proposed to function as an oncogene or as a tumour suppressor depending on the cellular context.
There has also been a substantial update for VHL, a tumour suppressor gene that plays a role in a rare inherited disorder called Von Hippel-Lindau syndrome but also in sporadic forms of cancer. The current update in COSMIC brings together the historic collection and the latest published data on the somatic mutations in the VHL gene, including novel mutations and VHL mutations in new histological entities and ethnic groups.
We have also included the fusion pair ETV6-ABL1, resulting from t(9;12)(q34;p13) or a complex rearrangement, is a rare but recurrent fusion in a wide range of haematological malignancies including myelodysplastic neoplasm, acute lymphoblastic leukaemia, acute myeloid leukaemia and Philadelphia chromosome-negative chronic myeloid leukaemia. Two types of ETV6-ABL1 transcript are detected: type A has an ETV6 breakpoint at exon 4 and type B at exon 5. The ABL1 breakpoint is consistent at exon 2. Both types result in constitutive tyrosine kinase activity similar to that seen with the BCR-ABL1 fusion. Eosinophilia is a common characteristic of patients with ETV6-ABL1 fusion.
The drug resistance pages have been updated with new samples and resistance mutations related to EGFR and ALK.
The changes to the Cancer Gene Census are now reflected on the website, and genes can be filtered by either tier 1 or 2 or both. There are now an additional 5 genes in tier 1 and 127 genes in tier 2. There has also been another update to the Hallmarks of Cancer feature to include both tier 1 and 2 genes. Full details are explained here.
Further website updates include being able to switch between the GRCh37 and GRCh38 reference genomes on all the main pages, see here for further details.
Finally for technical details please see the release notes.