
Cancer Gene Census, Hallmarks and the new tier system
3 Aug 2017
The Cancer Gene Census has had a thorough re-evaluation as part of integrating the Hallmarks of Cancer feature. This feature, initially released in May, has now had a substantial update to include 226 genes. It comprises of manually curated functional descriptions of genes and describes their role in developing the Hallmarks of Cancer as defined by D. Hanahan and R. A. Weinberg. This has been undertaken in collaboration with Open Targets, utilising the resolution provided by manual curation, each genes role in cancer is described based on the published scientific evidence. We did a full write up on the Hallmarks feature here.
In order to provide high-confidence and comprehensive data, new and more stringent criteria based on the level of evidence supporting each gene as a driver in cancer has been applied to the Cancer Gene Census. As a result, the Cancer Gene Census has been split into two tiers.
We now have a gold standard, tier 1, where a gene must possess documented evidence of activity that may drive cancer, as well as evidence that changes to protein activity promote oncogenic transformation. We also take into account the existence of somatic mutation patterns in cancer samples typical for tumour suppressor genes (usually characterised by a broad range of inactivating mutations) or oncogenes (usually characterised by well-defined hotspots of missense mutations).
Tier 2 contains genes that have strong indications of playing a role in cancer but with less expansive evidence than tier 1, it currently contains 41 genes however these are not reflected in the website or downloads due to data constraints with this release. The current Cancer Gene Census in COSMIC just contains tier 1 genes, however, as tier 2 is being expanded, with an initial planned release of about 200 genes in November 2017.
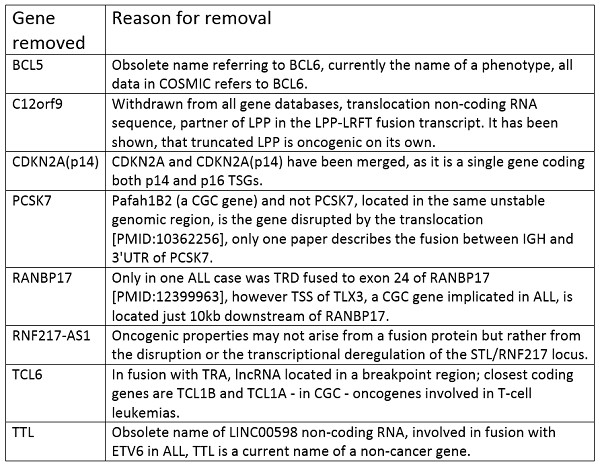
Finally, for various reasons, 8 genes have been entirely removed from the Cancer Gene Census. Full details of which genes have been moved to tier 2 and which genes have been removed entirely is given in the tables below.
The Cancer Gene Census genes that have moved to Tier 2, fall into six categories relating to why they have been moved to tier 2, these are fully explained below.
1. PMS1 - a component of DDR, only one recurrent frameshift mutation K164fs*6 in four samples, newer papers about MMR genes in cancer don't mention this gene, mice deficient in PMS1 do not develop tumours, no evidence for significant activity in MMR in vitro [PMID: 10542278]
2. Fusion genes with only one case (or rare partners of potent
oncogenes known to be fused to multiple partners and able to drive the
transformation on their own)
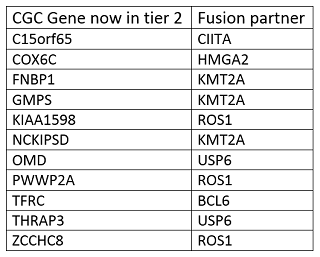
3. Fusion genes transcribed with a shifted reading frame or untranscribed upon fusion, for which there is no sufficient evidence for tumour suppressing activity.

4. Non-coding genes and pseudogenes do not fit to the current schema of Tier 1 of the Cancer Gene Census. We are working on better characterisation of the role of such genes in cancer. Temporarily they are classified as Tier 2 CGC genes.

5. Genes known to be involved in cancer only through fusions, where the oncogenic mechanisms depend on disruption of the structure of their fusion partner and there is no evidence of their other cancer-promoting activity so far.

6. Genes known to be involved in cancer only through fusions, for which there is not enough data describing their participation in oncogenic transformation.
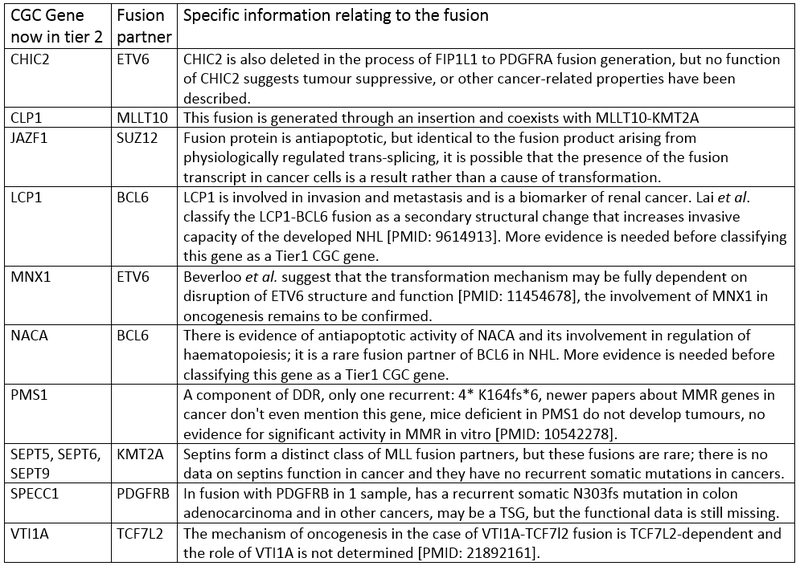
Finally the genes that have been removed entirely from the Cancer Gene Census
Currently only the tier 1 genes are shown in the Cancer Gene Census. The first 200 tier 2 genes will be released in November 2017, along with COSMIC v83. If you have any comments or concerns about the Cancer Gene Census or the Hallmarks features then please let us know at cosmic@sanger.ac.uk.
