Where has my mutation gone?
20 Jul 2017
It is the nature of maintaining a large database that there is a degree of turnover as mutations are reclassified and new information comes to light. As a result of our continuous efforts to ensure we provide the latest and most accurate information, we've recently received a number of enquiries regarding ‘missing’ mutations. Now that we have a blog, it seemed the perfect place to offer a clear explanation of why, in a few cases, mutations which existed in previous versions of COSMIC have been removed from the current version.
COSMIC contains millions of mutations, each with a unique ID which is stable across releases. However, over the years several mutations have become deprecated or merged with other mutations, which is why they appear to be missing from more recent versions of COSMIC. Below we have outlined the main reasons why a mutation may be removed:
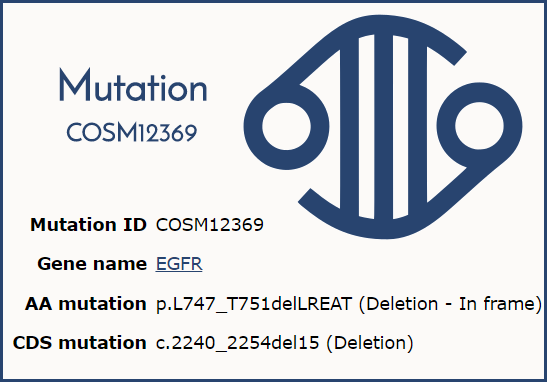
As part of recent integrity checks on the data, any duplicate mutations have been merged. How do the duplicates occur? Usually because of how they are reported in the literature. One example is the EGFR mutations COSM6254 (c.2239_2253del15, p.L747_T751delLREAT) and COSM12369 (c.2240_2254del15, p.L747_T751delLREAT). Here, the deletion was reported in different publications with different CDS locations so both mutations were entered into COSMIC, however, the resulting mutation is the same. Therefore, following the HGVS guidelines which indicate the correct syntax uses the 3' most deletion, COSM6254 has been merged with COSM12369.
Occasionally a reported mutation is included in COSMIC but later evidence confirms the variant to be a SNP, so the original mutation is therefore removed from the website, but is still available in the download files.
As part of ongoing syntax checks and updates in order to bring all the mutations in COSMIC up to date with the current HGVS syntax, occasionally mutations have been found to be completely duplicated. These are then merged to just one mutation ID, we are working to provide a list of which mutation has been merged into which and should be able to provide you with this shortly.
Data analysis in the field of genomics is constantly evolving, and analysis algorithms have changed and improved greatly over the lifetime of COSMIC. With this, the same data is occasionally reanalysed with slightly different outcomes and thus changes to the mutations reported. As some of the data in COSMIC derives from the International Cancer Genome Consortium (ICGC) who carefully recompile all of their data before each new release, these changes can often be reflected in their data sets; whilst COSMIC does not implement all the changes that ICGC make, occasionally legitimate data contained in a prior release may not appear in a current release.
Very occasionally a mutation is discovered to be incorrect and is therefore removed.
Unfortunately, at this point we do not have a way of tracking the lifecycle of individual mutations in COSMIC, however we are looking at the feasibility of implementing such a system and will keep you updated on progress. If you have any further queries then please contact us at cosmic@sanger.ac.uk.
