Update to TSC1 and TSC2 genes
19 Mar 2019
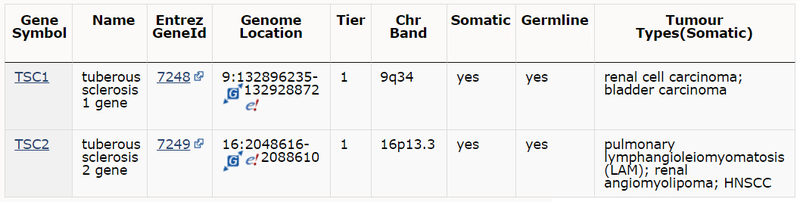
As part of release v88 we have focused on updating the expert-curated mutation data for TSC1 and TSC2 genes. This includes approximately 60 additional publications that include mutation screening data for these genes.
Tuberous sclerosis complex (TSC) is an autosomal dominant disorder characterized by skin manifestations and formation of multiple benign and/or malignant tumours in different organs. In addition, TSC disease causes disabling neurologic disorders, including autism, intellectual impairment and seizures. Postzygotic somatic mutations in TSC1 or TSC2 result in mosaic forms of tuberous sclerosis complex, which may present as disseminated or segmental disease, or with mild disease or later onset than the inherited disease.
TSC1 and TSC2 encode proteins hamartin and tuberin, respectively, which interact and form a heterodimer that inhibits the activation of mammalian target of rapamycin (mTOR) complex 1, a master regulator of nutrient and growth factor induced signalling. As common for tumour suppressors, tumorigenesis involves inactivation of the second allele, typically via a second-hit somatic mutation in the wild-type allele of the TSC gene. Sporadic tumours involving somatic mutations in TSC1 or TSC2 with no apparent germline mutation are also found. Current COSMIC release reports somatic mutations in renal cell carcinoma (RCC), subependymal giant cell astrocytomas and other gliomas, bladder cancer, hepatocellular carcinoma, melanoma, angiomyolipoma and lymphangioleiomyomatosis amongst many other tumour types.
In a study of 91 mucosal melanoma patients the overall somatic mutation frequency of TSC1 was found to be 17.6%. The TSC1 mutations were mainly missense mutations and were found across 11 different exons. The mutations were more inclined to occur in advanced mucosal melanoma and these patients had a worse outcome than patients without TSC1 mutations (COSP44621).
In a case of a young adult with renal epithelioid angiomyolipomas (EAML), a rare tumour type with aggressive behaviour, a complete and durable response to sirolimus was achieved, the patient being disease free after 36 months of treatment (COSP45400). Targeted next generation sequencing (NGS) of MTOR, TSC1 and TSC2 genes revealed one inactivating TSC2 mutation (c.2739dup; p.K914*) in the tumour cells. The authors demonstrated mTOR pathway activation and TSC2 inactivation as the mechanism for the response.
Similar examples of significant clinical response to mTOR inhibitors in other patients with sporadic or inherited disease such as RCC and Hodgkin’s lymphoma have also been described, and some of these publications are included in the current release. These studies support sequencing as a useful tool to identify patients sensitive to mTOR inhibitors and support mTOR inhibition as an important therapeutic approach in these malignancies.
