“Subtraction Analysis”: COSMIC’s role in defining causes and consequences of clonal haematopoiesis
23 Jan 2023
Prefer to listen to the interview? Head over to our brand new podcast 'Conversations with COSMIC' to hear the full interview with Siddhartha and follow us for more episodes coming soon!
Siddhartha Kar is UKRI Future Leaders Fellow at the MRC Integrative Epidemiology Unit at the University of Bristol. He initially trained as a medical doctor in India, before moving to study public health and epidemiology in the United States. Siddhartha completed his formal education with a PhD in genetic epidemiology at the University of Cambridge in 2017. He’s held his current post since 2020 and humbly described it as ‘running a small computational lab in Bristol’, although speaking to him it is much more than that.
We caught up with him to learn more about his recent publication 'Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis', and how COSMIC was involved in the research.

Can you summarise your research and recent publication in one sentence?
In terms of publications, this is the largest and most comprehensive analysis of inherited genetic factors that confer risk of a condition we call clonal haematopoiesis (CH), and the downstream consequences of CH.
Can you please briefly describe what you mean by clonal haematopoiesis?
Clonal haematopoiesis is not a disease, it's a phenomenon that's quite common and is related to ageing. There are two components to it. Haematopoetic stem cells, or HSCs are the professional blood making cells - there are about 10 to 20,000 of them . When these stem cells or their ‘children’ (progenitors), acquire mutations in their DNA that give them some sort of evolutionary or fitness advantage, they start multiplying and forming a clonal population that differs from the ‘normal’ blood cell population. You can think of it like Darwinian theory, the cells that have the mutation have an advantage and form a new sub-population.
And it’s literally in the name. Clones because they’re all replicas of the one original cell with the mutation, ‘heme’ is the Latin root for blood, and ‘poesis’ derives from Ancient Greek meaning the process of bringing something to life that didn’t exist before.
This phenomenon was identified in the 1990s. But it's only in the last 10 years that we've had the technology come in on the sequencing and computational side to be able to detect it at large scale.
Do these clones behave abnormally or do they do their normal functions?
It's actually not very well understood. We know that having these mutations allows the clone to replicate, giving it an advantage in line with Darwinian evolution’s ‘survival of the fittest’. So with this inherent fitness, it starts to become about 2% of all your blood cells. And in someone with acute myeloid leukaemia, almost 100% might be dominated by the clone. But does that prevent the cloned type of blood cell from doing its normal activities? We don't think so. It's just that cells and organisms have this inherent drive to grow and create a next generation.
And how common is this phenomenon?
It tracks and increases with age. Below the age of 40 years, less than 1% of individuals usually have it, but by the time someone reaches 70, 10 to 20% of individuals would have it.
What’s the background to this research and why were you interested in it?
I've talked about what the phenomenon is, but why is it important? And that's where the clinical background comes in. This phenomenon is associated very deeply with blood cancer risk. It's been observed over the last 10-15 years that those with CH have over tenfold higher risk of developing a blood cancer.
I've spent the better part of the last 10 years studying the epidemiology and genetics of non-blood cancers. And the process of CH probably occurs in other tissues as well, in the breast, in the lung, in the colon. But it's very difficult to get tissue on a very large scale other than blood, the only other comparable tissue is the buccal or oral mucosa, the sort of thing that is taken during a COVID swab. Since we can't get such large-scale data on other tissues, I got very interested in the process of CH, and using the understanding to inform other types of cancer.
In addition, a series of high-profile publications over the last 10 years have associated CH with a bunch of other diseases, most notably cardiovascular diseases. For example, coronary artery disease, which includes things like heart attack and angina, and stroke, particularly ischemic stroke. But these were relatively smaller studies. The link with blood cancer seems intuitive, and we’re curious to find out: are the other diseases that link with CH causally associated? Or is it just that ageing causes some of these other diseases and ageing also causes clonal haematopoiesis?
So what were you hoping to learn in this research? What were your aims when you set out?
So far only four genetic variants have been found to be associated with CH. These variants are inherited from parents and have an impact on what happens over the course of our life. Firstly, we hoped to find new inherited genetic regions or variants that confer risk of clonal haematopoiesis. After identifying these regions, we planned to look at nearby genes to learn about the biological pathways that lead to this.
Secondly, we planned to use a really powerful tool called Mendelian randomization to identify the causes of CH, consequences of CH, and potential prevention/treatment strategies.
Before leaping into results, let's discuss some of the methodology exactly how you went about studying it. In particular, where did COSMIC add value?
The main study is what we call a genome wide association study, or a GWAS, which we conducted on 200,453 individuals from the UK Biobank. This is basically a case-control study where we compared the inherited genome of those who have CH with those who don't, to ask whether certain variants were more common in CH.
Before comparing, we had to define the ‘CH phenotype’ based on bloods and exome sequencing data. CH is defined based on the presence of specific mutations in one of 43 genes (a small number of individuals have it in more than one but that is relatively rare).
But how do you know whether these mutations are inherited or acquired? A mistake at this point would result in our entire analysis falling apart.
And that’s the value of tools like COSMIC. For a researcher like me and other members of my team, COSMIC provides an authoritative catalogue of common mutations in the somatic genome. We can trust the source of mutations, mutational signatures, copy number signatures, and cell line-based data. It enables us to do a 'subtraction analysis' - which is not an official term, I'm coining it now! More specifically, we used COSMIC to filter the mutations that occur in the somatic genome in blood cancers. If we saw a particular mutation in one of these 43 genes occurring in blood cancers in at least seven COSMIC records, then we determined that this must be a somatic mutation acquired over the lifetime rather than inherited. And that – among other filters – helped us define this phenotype – the outcome of CH.
You also looked at different ‘subtypes’ of CH - can you explain that more?
We studied CH as one global entity, incorporating all 43 genes, but we also looked at five individual subtypes. Firstly, we looked at the most commonly mutated CH genes (out of the 43) separately, DNMT3A and TET2. More than 50% of CH cases have somatically acquired mutations in DNMT3A. It’s a gene involved in epigenetic regulation and has links to acute myeloid leukaemia (AML), which is recorded in COSMIC.
We also considered the size of the clones. So if a clone has taken over less than 10% of the blood, that's a small clone, if it occurs in more than 10% of the blood, that's a large clone.
So our subtypes were; Global, DNMT3A-mutant, TET2-mutant, Small Clone, and Large Clone.
You briefly mentioned a technique called Mendelian randomisation?
When you have GWAS data, one of the most powerful things you can do is apply a technique called Mendelian randomisation.
So let's look using smoking as an example. With traditional methods, we would ask people if they smoked, then compare the smokers and non-smokers for particular health outcomes. But, people might not fully report their smoking behaviours or they might develop a disease and then stop smoking. And this can mix up the findings - it would 'look' like low smoking is protective for the disease but it’s actually reverse cause and effect. These problems are referred to as confounding and they might dilute the association between smoking and a particular disease outcome.
So where does Mendelian randomisation come in? It's like we've all been selected into nature's own randomised trials as inherited variants are randomly determined and fixed at the point of conception. That provides an interesting natural experiment because some variants are known to be robustly associated with particular behaviours. With regard to smoking, there are 380 variants known to increase a person's risk of acquiring smoking at some point of time in their lives, usually early in life. So you can then use those weightings to divide your population into those with the variants and those without as being those at higher/lower risk of smoking, then compare health outcomes in these two groups. This removes the dependency on the individuals reporting, or the disease itself affecting, smoking behaviour. This enables causal inference, making it easier to determine that smoking does cause this particular outcome.
To note, the risk factors are not deterministic, it doesn't mean that somebody who has these will go on to smoke. But there's enough of a propensity to consider the impact of the variant. The common example is variants in the gene CHRNA5 which is one of the first smoking related genes. This gene encodes a key part of the nicotine receptor. So some of us have a nicotine receptor that responds to the nicotine in a way that we feel very good about smoking very quickly and progressively need to smoke more to derive the same ‘feel good’ feeling. Others don't. And that's why some people take to smoking, enjoy it and continue smoking, whereas others don't.
We use this technique as it’s a clever way of leveraging all the genetic data to answer questions that are fundamentally causal and deriving answers that allow us to actually intervene, because getting someone to stop smoking is a very constructive public health intervention.
So we started with GWAS, then heterogeneity of the phenotype ‘clonal haematopoiesis’, and finally Mendelian Randomisation for causes and consequences.
In terms of looking across diseases, rather than just looking at cancer, do you think we should be doing more of that in research? Do you think there's more overlap than we have typically understood before now?
Yeah, there's definitely more overlap. I wouldn't blame those who looked at this before for studying single disease contexts, because datasets with linkage across entire lifetime medical records and diseases encountered weren’t available. It's only in the last 10 years or so that resources like the UK Biobank have become mature. They have the necessary size and linkage with medical records to tell you about the whole spectrum of diseases which makes it easier for researchers like us to ask questions across multiple different disease areas.
Onto the exciting bit - can you talk through the key findings?
Yes, I’ll break it into two broad sections in line with our hypothesis.
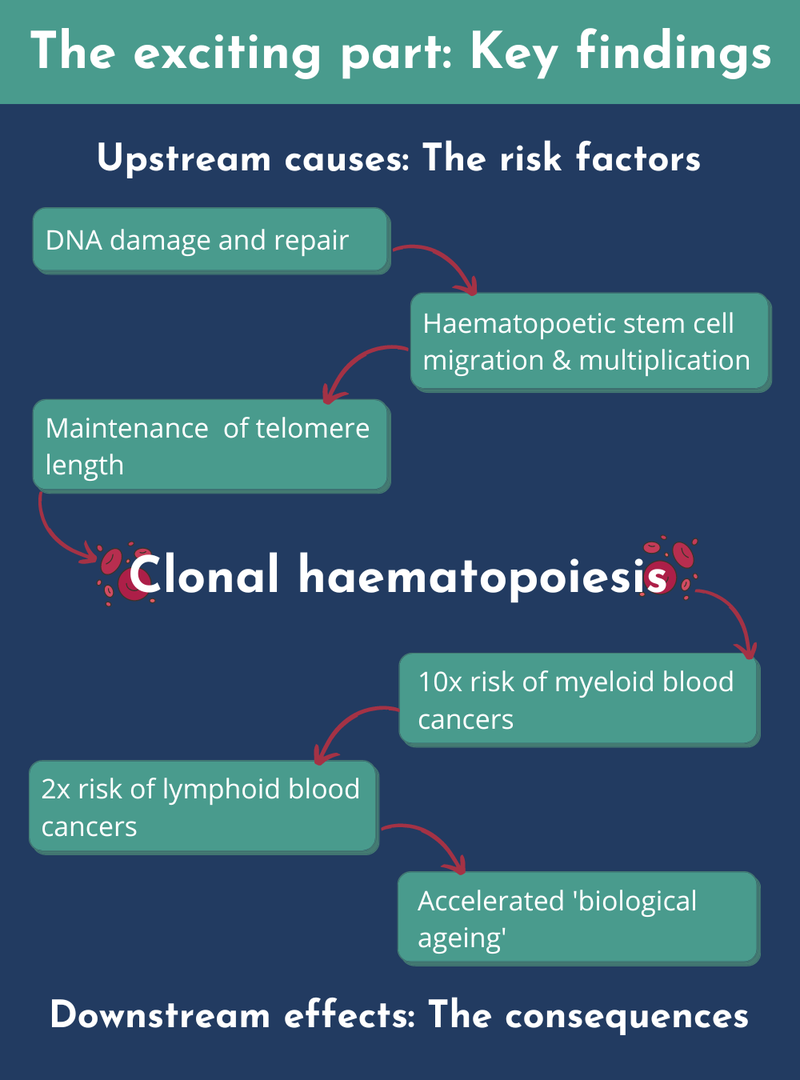
The key finding is that we went from four to 14 genetic inherited risk factors for CH. We associated these genetic risk factors with three processes - this gives us clues to the pathways that biologically lead to CH. Firstly, and this is the key one, is DNA damage and its repair. Second is the way haematopoietic stem cells migrate and move around as they multiply. Third is the maintenance of telomere length.
Telomeres are ends of the chromosomes and the longer ones tend to confer higher risk of developing CH. Why do they affect risk? Partly because there's a biological maintenance mechanism for these telomere lengths that’s impacted by these inherited variants.
In terms of the causes of CH, I’ve mentioned telomere length but the biggest modifiable cause is smoking. Both of these extended across all the types of clones, and both major genes that I talked about, DNMT3A and TET2, which are somatically mutated. We found two significant factors associated with this. To a lesser extent, larger clones - the ones making up more than 10% of the blood population - are associated with higher body mass index (BMI). All these findings were uncovered by our Mendelian randomisation analyses.
We found that CH confers more than a ten-fold risk of myeloid blood cancers, and about two-fold risk of lymphoid blood cancers. We looked at whether the cancer was driven by smoking, but the CH downstream effects are independent of smoking.
We didn't see any evidence that CH causes cardiovascular disease using the Mendelian randomisation approach, with the exception of atrial fibrillation - an irregular heartbeat. And the final thing that we found was a risk developing epigenetic ageing. Our epigenomes tend to have changes during the lifespan, and some relate to the discrepancy between ‘chronological age’ versus ‘biological age’. We found that having these mutations accelerated someone's ‘biological ageing’.
And what about links with any other cancer types like you mentioned in the original hypothesis?
Yes, so the link with blood cancers acted almost like a positive control - if it didn't show up, we would have thought something was terribly wrong with our analytical methods. But we were surprised to find that CH was associated with higher risk of prostate cancer, lung cancer, oral cancer, and ovarian cancer. To note, we do not think that CH is directly causing these, more that the process happening in the blood is mirroring a parallel but very similar process occurring in the lungs, ovary, and prostate. We can't prove this, but these findings indicate the possibility that CH is a marker of parallel processes involving other genes, which are getting somatically mutated and causing these cancers in other parts of the body. It’s relatively easy to acquire and study 200,000 blood samples to get the scale required - it would be quite tricky to get that many prostate samples.
Now, I want to quickly touch on Ancestry because it is mentioned in the paper. In this paper, you've discussed European ancestry. Why is it important to consider ancestry?
Absolutely - I'm very interested in the ancestry question myself. As you know, I am from India, and a relative minority in the UK!
The answer to your question is a bit nuanced. Why did we study European ancestry? One of the reasons is that the UK Biobank has an overwhelming European ancestry. And even then we could have broken it down further because Scandinavians are different from Italians.
But the other reason we didn't really look at other ancestries is about keeping it simple for ‘proof of principle’. As a relatively first-of-its-kind study, we needed to be certain that we were picking up the inherited differences that associate with the condition and not inherited differences between individuals of different ancestries. It’s why we study homogeneous sets of people in some of these analyses like smokers only, or non-smokers only.
The second aspect of ancestry is to ask, 'does clonal haematopoiesis itself differ across ancestries?'. And there is some evidence out there to say that this does differ. Taking this work further, I’d like to do similar genetic analysis in cohorts which are enriched for British South Asians (as an example). In the UK, we now have a number of cohorts where the focus is on non-European ancestry - and a couple of them are ‘East London Genes and Health’, and ‘Born in Bradford’. And of course, there's a lot that needs to be done elsewhere in the world - there's actually already data on CH from the Japan Biobank.
How do you hope other people will use these findings? Are there any clinical implications?
The medic in me will say that we have to be very cautious in jumping from basic research published in Nature Genetics to immediate clinical applications, and there are a lot of steps down to translation. But with that disclaimer, there are two interesting insights.
Firstly, we found an inherited variant in the PARP1 gene that affects someone's risk of acquiring the ‘DNMT3A-mutant CH’ . The product of this gene is the target of PARP-inhibitors, which are currently used for treatment of ovarian and breast cancers. This opens up a whole range of research questions: could PARP-inhibitors prevent the acquisition of ‘DNMT3A-mutant CH’? Does PARP inhibition therefore have a role in preventing blood cancer? It's very exciting that we've seen variants in a gene with a drug already in the clinic, and the next step could be to launch small trials to look at whether this works in a real world setting.
Secondly, as we increase our understanding about the inherited genetic risk factors, we're able to bring these together to create polygenic risk scores. These would allow us to predict who's going to develop clonal haematopoiesis. If that’s taken further, we could combine the data more robustly to predict who's most likely to develop leukaemia. So we hope it will help in risk stratification and early detection of cancers - in particular leukaemia.
In the very long term, with much larger samples and refinement, we may be able to answer whether this is useful in the detection of other cancers marked by similar clonal expansion processes. But that's deep into the future. I might be quite old by the time that happens!
What's next for you?
As discussed above, I’m interested in the therapeutic development and risk prediction aspects of this. And for me, there’s also a third avenue - most of the variants actually lie in bits of DNA that used to be called junk DNA, or non-coding parts of the genome. The statistics tell us that they increase risk, but we don’t know exactly how that happens. We hypothesise that they’re acting on pathways, and our research hints at DNA damage and repair mechanisms. This is fertile soil for further research, and once the pathways are understood, it's possible to manufacture drugs that intervene on that pathway, which might prevent multiple cancers.
Will you continue to use COSMIC in this research?
The thing with COSMIC is that it's so ubiquitous, you don't realise that you're using it until someone actually asks you like this. So in this study, we studied those who don't have cancer. But we’re doing other studies looking at the tumour genome and asking a similar question - does the inherited genome increase my risk of having a particular mutation in the tumour? Because there's a vast expanse of mutations in the tumour, we need to single out passenger versus driver mutations. That's where resources like COSMIC come in, because it’s continuously expanding and understanding the state of the art in terms of what the drivers are.
We are already using COSMIC’s copy number signatures in the work mentioned. But another study we are hoping to do in the future is to potentially identify mutational signatures of CH in the whole genome sequencing data. This study was based on exome sequencing, which is not very reliable for identifying mutational signatures. But UK Biobank has already released whole genome sequencing data from 300,000 individuals. And by next year, all 500,000 individuals will have their sequence data released. As we move from single genes to looking at mutational signatures of CH, COSMIC mutational signatures will help us to identify the robustly known ones.
How much of this is a collaborative effort?
Oh, it relied heavily – and I can’t stress this enough – on collaboration and interdisciplinarity. I was working with George Vassiliou who is a practicing haematologist, and members of his team who have deep bioinformatics expertise, in particular, Pedro Quiros. George, Pedro and I co-led the work. The interdisciplinarity is one of the most enjoyable aspects of this paper. It has been immensely rewarding. Each of us have brought different perspectives to the research question. For instance, George was interested in understanding the risks, asking questions like, ‘How do I counsel my patients?’ Do I tell them that they are at risk for this vast majority of diseases because of clonal haematopoiesis? Or do I tell them to watch out for blood cancers?’
And now we can give better answers that there's tenfold increased risk of blood cancers, but the increased risk of heart disease from the common, global form of CH is perhaps not as important.
How did you celebrate the paper?
Yes – well speaking of the wider team. Because of the pandemic we hadn’t actually met in person until very recently in Cambridge, and that was to celebrate the paper. We uncorked two bottles of champagne. This entire paper was done during some of the early lockdowns and subsequently online. And so it was not just a celebration but a good laugh that we managed to pull it off, working 18 months together without actually meeting!
