Mutational Signatures Update
15 May 2019
Somatic mutations are present in all cells of the human body and occur throughout life. They are the consequence of multiple mutational processes, including the intrinsic, slight infidelity of the DNA replication machinery, exogenous or endogenous mutagen exposures, enzymatic modification of DNA, and defective DNA repair. Different mutational processes generate unique combinations of mutation types, termed “Mutational Signatures”.
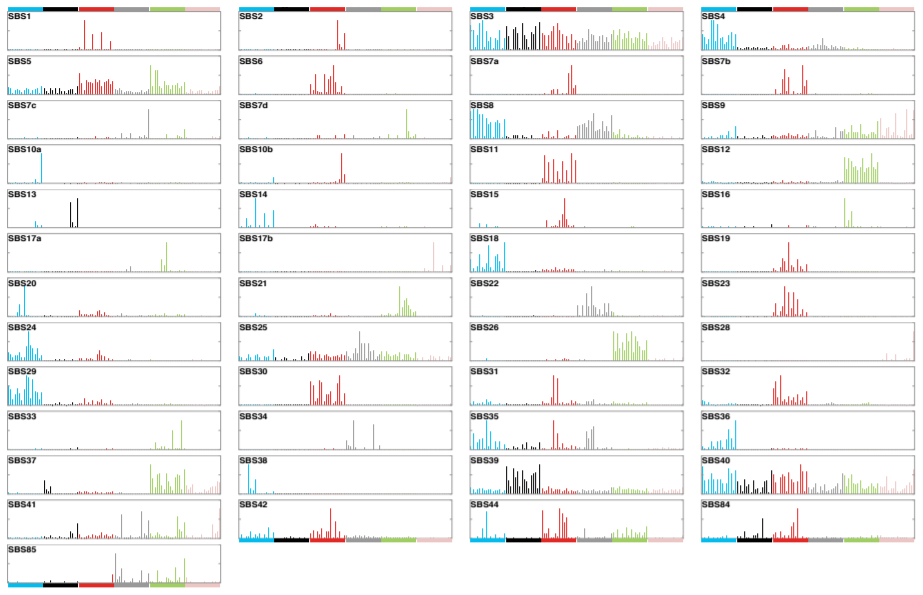
Earlier studies (Nik-Zainal S. et al., Cell (2012); Alexandrov L.B. et al., Cell Reports (2013); Alexandrov L.B. et al.,Nature (2013); Helleday T. et al., Nat Rev Genet (2014); Alexandrov L.B. and Stratton M.R., Curr Opin Genet Dev (2014)) identified 30 mutational signatures. These were curated to form a census of signatures and have been shown on the COSMIC website since 2015. We are pleased to announce a major expansion of the mutational signatures, based on the analysis of 84,729,690 somatic mutations from 4,645 whole cancer genome and 19,184 exome sequences encompassing most cancer types, we recently characterised 49 single base substitution, 11 doublet base substitution, four clustered base substitution, and 17 small insertion and deletion mutational signatures.
In this release of COSMIC mutational signatures, we provide a summary “vignette” for each mutational signature. The vignettes include mutational profiles and notes on potential aetiology, associated mutational signatures, how the signature has changed during iterations of analysis, and other pertinent comments. These are intended to be short, very high level, “aides-memoire” for key elements of what we know, suspect or has been widely discussed for each signature. They are not intended to be comprehensive summaries of everything reported in the scientific literature and they are not referenced. Further, we provide access to all input data and references for original sources that were used to derive these mutational signatures (Alexandrov L. et al. 2018).
The final sets of reference mutational signatures were determined from the ICGC/TCGA Pan Cancer Analysis of Whole Genomes (PCAWG) Network analysis, supplemented by additional signatures from the other datasets. Signatures were supported by the outcomes of analyses using the 192 and 1536 mutation classifications, the existence of individual cancer samples dominated by a particular signature, and, where available, prior experimental evidence for certain mutational signatures (full methods).
Each signature has been allocated a number consistent with, and extending, the previous COSMIC mutational signatures. Some previous signatures have been split into multiple constituent signatures and these were numbered as before but with additional letter suffixes (eg, signature SBS17 has been split into signatures SBS17a and SBS17b). DNA sequencing and analysis artefacts also generate mutational signatures, and we indicate which signatures are possible artefacts these have also been included.
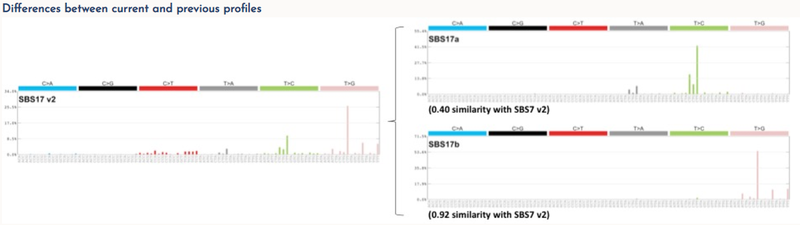
Both the expanded mutational signatures (v3) and the previous mutational signatures (v2) are available. Full details of this work are available in Alexandrov L. et al. 2018.
