In the literature
13 Nov 2018
The COSMIC team have been busy writing papers and we have published 3 articles over the last couple of months. The first, in Nature Genetics looks at our COSMIC-3D platform; the second, in Nature Reviews Cancer discusses the Cancer Gene Census; finally we have published a more general overview of COSMIC developments in the NAR database issue.
The COSMIC-3D platform, developed in conjunction with Astex Pharmaceuticals can been used to explore cancer mutations in the context of three dimensional protein structures. It allows for high-resolution interrogation of the structural and functional nature of cancer mutations allowing for the identification and characterisation of drug targets, through the use of an interactive website.
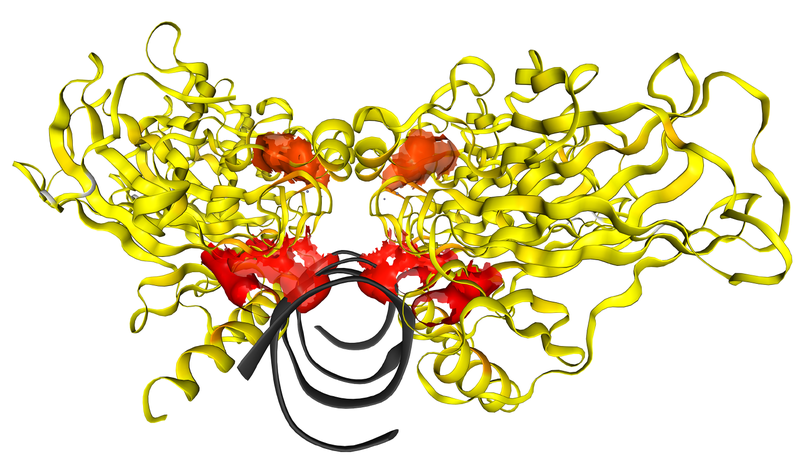
Fig. 1 A screen capture of TP53 showing the core domain binding DNA.
The recurrence of missense cancer mutations across protein structures can be viewed for each individual gene. Individual mutations can further be selected from a sequence viewer, and displayed in the context of 3D protein structures. Understanding the position of mutations across experimental protein structures, and their juxtapositions with known and predicted small-molecule binding sites also provided in COSMIC-3D, paves the way for using mutation data to generate hypotheses of the impacts of mutations on protein structure and function, and for "Mutation-Guided Drug Design" in precision oncology. To learn more, please take a look at the article, available from Nature Genetics. Alternatively the full-text of the article is viewable without a Nature Genetics subscription thanks to the Springer Nature SharedIt initiative.
The Cancer Gene Census provides a comprehensive summary of over 700 genes known to be strongly implicated in causing human cancer. The census also provides functional descriptions of the changes that occur to drive the disease in terms of the ten Cancer Hallmarks - biological processes that drive cancer.
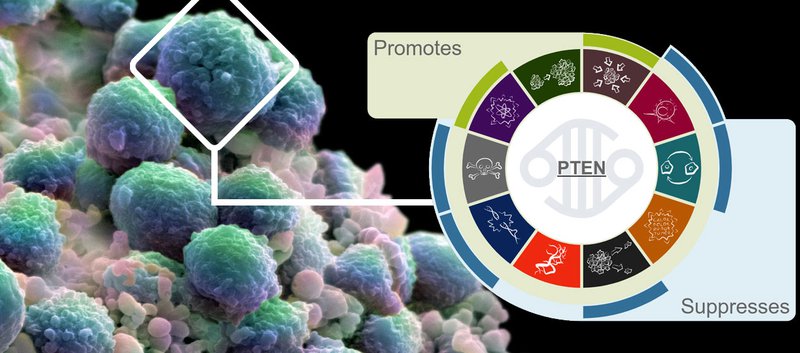
Fig.2 Summarising the Hallmark behaviours of PTEN in cancergenesis.
The scientists have manually condensed almost two thousand papers to draw together strong evidence of each gene’s role in cancer, and describe which genetic functions go wrong in the process. The full Cancer Hallmarks can be explored interactively on the COSMIC website, with clear concise explanations of the behaviours observed in each gene. To read more, please see the full article available from Nature Reviews Cancer.
Finally, we have written a broad overview of the developments in COSMIC over the last eighteen months, this has looked at both new data and changes to the website and data access. One of our curators has also outlined how the expert manual curation is carried out within the COSMIC team. This gives an insight into both how thorough and technical the curation is and also provides transparency to the process. This was published at the end of October and is available to read from Nucleic Acids Research.
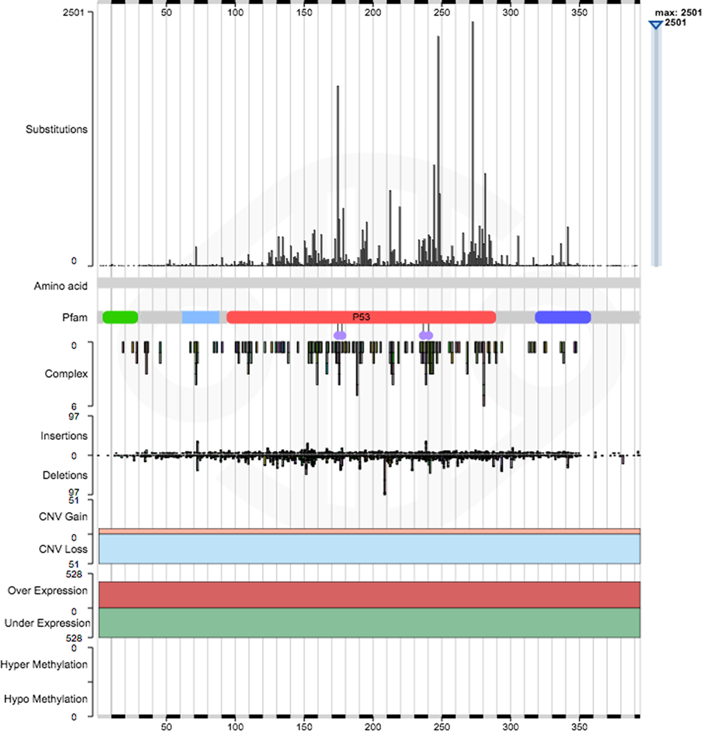
Fig 3. The gene histogram for the tumour suppressor gene TP53, showing the range of data available for TP53.
We hope that reading these articles provides some perspective on how COSMIC is developing and adapting to the changing landscape of cancer genomics. If you have any questions or ideas please get in touch cosmic@sanger.ac.uk.
