Drug Resistance data in COSMIC
6 Dec 2017
Drug resistance is inherently complex, both in its mechanism of action and how it is measured and reported in the literature. With the increasing impact of precision medicine, scientific literature is emerging which describes genetic responses to targeted therapies. Resistance to particular drugs may occur in some patients following an initial response, and this is often due to newly acquired mutations.
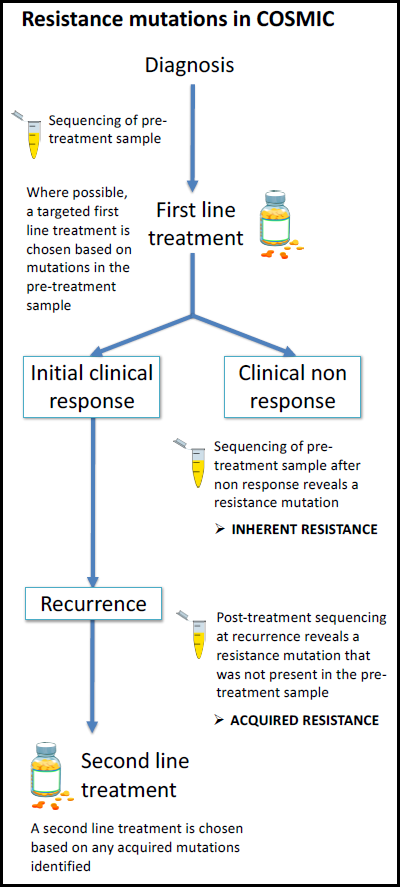
Since 2016, genetic curation in COSMIC has been extended to include both acquired resistance mutations and their effects on drug responses, as well as inherent (pre‐treatment) resistance mutations. Prioritising well‐characterised drugs and genes, COSMIC v83 contains resistance mutation profiles across 23 drugs, detailing the occurrence of 356 unique resistance alleles across 2016 tumour samples.
Within the COSMIC website, pie charts and histograms show the frequency of mutations in the evolution of therapeutic resistance, and additional mutational and clinical information associated with resistant samples can be explored. The full dataset can be downloaded from the COSMIC SFTP site.
Below, we describe in more detail how the different resistance data are classified and give a reminder of how they can be accessed through the website.
There are two ways the data can be accessed, firstly by clicking on the ‘drug resistance’ link in the data curation section at the bottom of the homepage. This brings you to an overview page that gives you a quick explanation and summaries the drug information currently in COSMIC, which is regularly updated.
Alternatively you can search directly for the gene you are interested in from the homepage and then navigate to the drug resistance tab on the gene page.
The drug resistance section will default to give you information on all the drugs that have been used to treat tumours with mutations in the gene you are looking at. It will also give you information on other genes that have resistance mutations to the selected drugs. For ease of understanding the graphs, it is simplest to limit the display to the drugs or genes you are interested in so that you only see information relevant to what you are investigating.
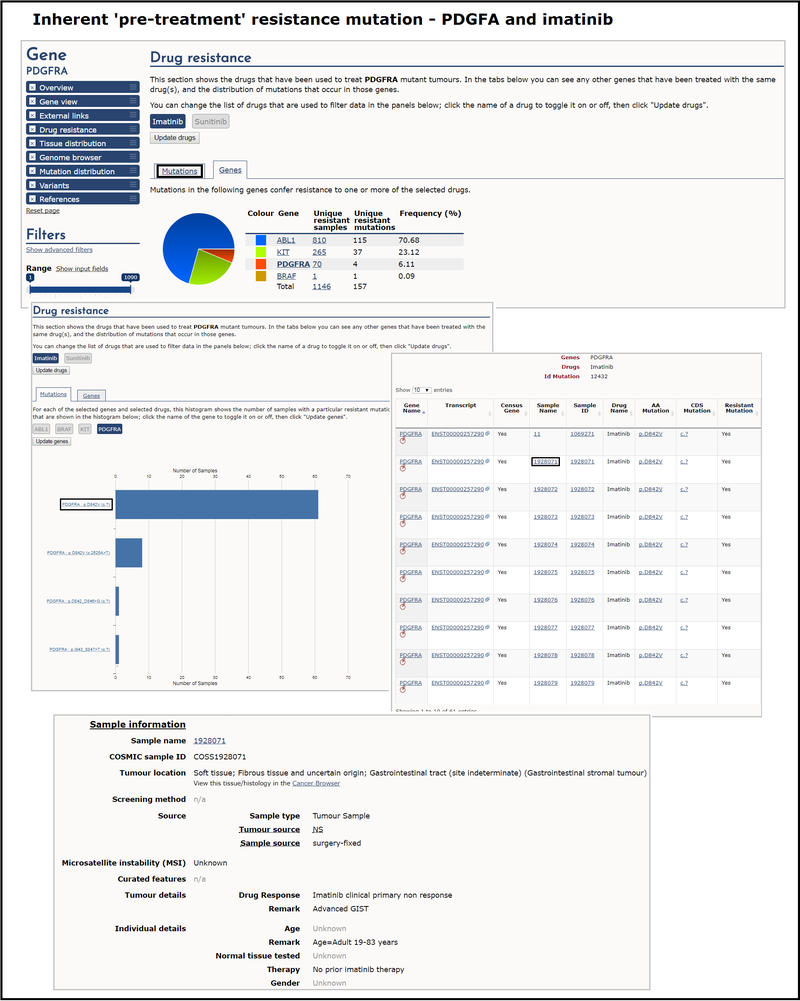
An example of inherent 'pre-treatment' resistance in COSMIC is the PDGFRA gene with the the tyrosine kinase inhibitor imatinib. The majority of samples from patients with gastrointestinal stromal tumours (GISTs) that contain PDGFRA p.D842V mutations exhibit resistance to treatment with imatinib. In order to investigate this, select the mutation tab under the drug resistance section of PDGFRA, de-select sunitinib to leave just imatinib active and click Update drugs. Then in the Mutations tab immediately below de-select all genes except PGDFRA and click Update genes. The resulting histogram shows the number of samples with a particular resistant mutation. Selecting the most frequent, p.D842V, brings up a table with all samples associated with that mutation, including transcript, site, AA and CDS mutation etc. You can click on individual samples for more information about the sample and drug response. This gives further details about a particular sample, including environmental factors such as patient age and prior therapies.
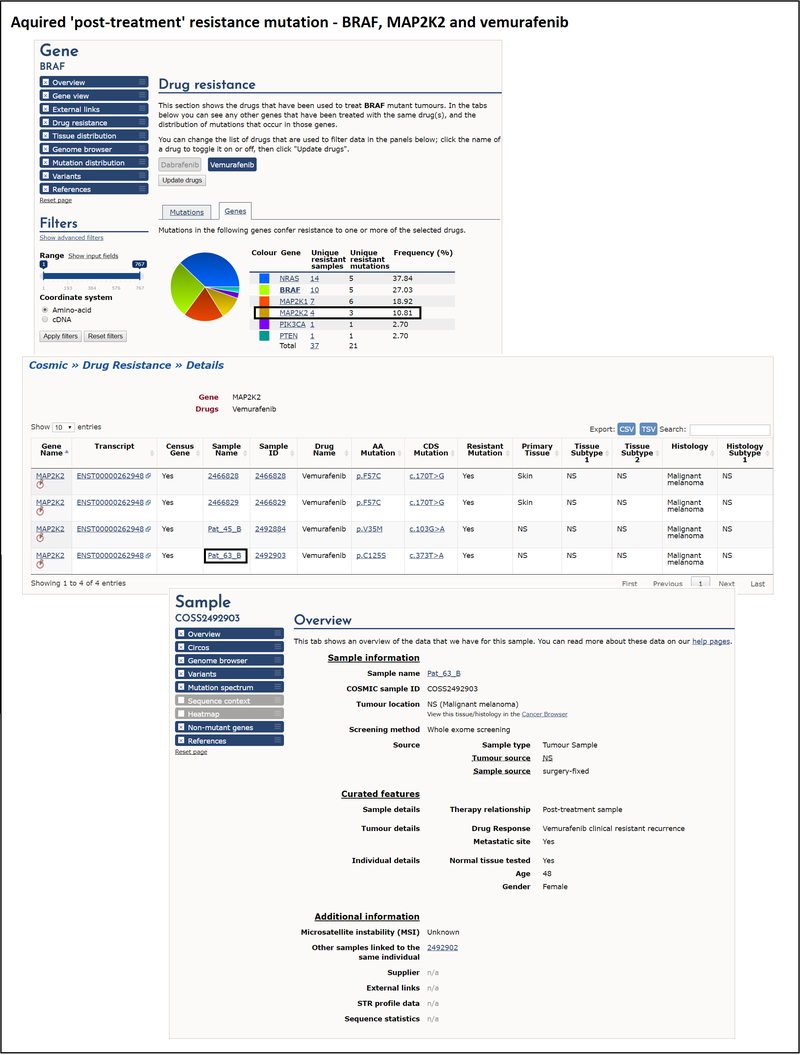
An example of acquired 'post-treatment' resistance is BRAF and the drug vemurafenib. Vemurafenib is a BRAF enzyme inhibitor that targets the BRAF V600E mutation. Patients with melanoma are tested for BRAF mutations to determine whether they are likely to respond to vemurafenib. After an initial response, some BRAF mutant melanoma patients relapse, and this is thought to be due to the acquisition of a second mutation, for example in the MAP2K2 gene.
Looking at the drug resistance tab on the BRAF gene page, and selecting vemurafenib as the drug, the genes table lists the genes that contain mutations conferring resistance to vemurafenib. Selecting the unique resistant samples column for MAP2K2 brings up a table where you can see all the vemurafenib-treated samples with mutations in that gene. Selecting a sample ID provides you with detailed information about that particular sample. This allows you to explore the clinical details associated with this sample, the presence of other variants and environmental factors and the reference from which this information was curated. Links to any other samples from the same patient are also shown under the additional information section.
In combination with improved molecular diagnostics, it is hoped that this new curation will further aid drug development and allow clinicians to better target therapies to individual patients. We will continue to add new drug/mutation combinations as new drugs are introduced and new resistance mechanisms identified. We are also looking at the possibility of curating additional data points that may better represent the therapeutic progress of individual patients with numerous treatment lines. Finally we are looking to introduce new ways to explore the drug resistance data and the possibility of showing aggregated results on the website. Feedback, particularly with regards to this last point would be most appreciated to cosmic@sanger.ac.uk.
