
Do you have data for COSMIC?
30 Jan 2018
COSMIC welcomes researchers submitting data to us directly either pre- or post-publication, the following article outlines how to do this.
Are your findings published? Firstly congratulations! In that case, we probably already know about your paper. As a matter of course we continually search the literature for papers relevant to both our currently curated genes and genes that are soon to be curated, so chances are your paper is already on the 'to do' list! However, you are more than welcome to drop us a line and check that we have seen your paper. Contact us on cosmic@sanger.ac.uk.
If you are pre-publication, we are also happy to accept your data (and generate a COSP identifier as a proof of data submission to COSMIC that you can communicate to a journal if required). Although at this point we would not be able to display your data on the website or in the download files until it can be linked to a peer-reviewed publication. Although we do periodically check, we also ask that you let us know as soon as your paper is published so that we can include your data in the next update of COSMIC.
What form should your data take? In order to make the curation process as efficient and accurate as possible, we have some guidelines about the format and level of detail that we require. By following these guidelines authors will contribute to the quick and efficient dissemination of their research results via COSMIC.
Samples:
- Data in COSMIC is curated per sample. Thus mutation or clinical data can only be entered if it is in reference to a sample. The minimum requirement for a sample to be included is the full mutation details at the nucleotide and ideally protein level (see below).
- Samples reported in an earlier publication will be excluded to avoid duplication. Indicating that a sample is a duplicate is very helpful.
- Papers reporting samples with more than one mutation (from one or more genes) please specify which mutations occur together in a sample.
Reference Sequences:
- Please state which reference sequence and version was used to describe the mutations (e.g. NM_0060165.3 or ENST00000215919). This makes mapping mutations to the COSMIC reference sequence much easier, and for some mutations is absolutely essential.
Mutations:
- Ideally mutations would be described both at the nucleotide and amino acid level. Please note we can not accept mutations
without genomic nucleotide coordinates.
- COSMIC mutation syntax is based on the HGVS syntax, it is useful if authors also use this nomenclature. For insertion mutations it is helpful if they are described as e.g. c.1118_1119insA rather than c.1118insA which can be ambiguous. If the protein result e.g. p.N373s*6 is also provided, the position can be confirmed.
Suggested presentation of results:
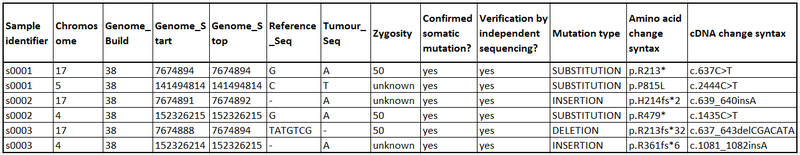
The table above shows a suggested layout which makes your results easier to incorporate into COSMIC. Clinical fields can be filled in if the data is available and additional columns can be added for further information e.g. smoking status, drug response etc. We have a template of this table available which can be adapted as necessary, alternatively we can also accept your VCF formatted files that you may already have. If you have any queries or would like to submit data, please get in touch at cosmic@sanger.ac.uk.
