
*NOW LIVE* Discover the The Cancer Mutation Census!
26 Aug 2020
What is The Cancer Mutation Census?
The Cancer Mutation Census (CMC) is an undertaking to classify coding mutations in COSMIC and identify variants driving different types of cancer.
We are often asked which are the mutations that matter the most - the CMC will help to answer this question! The CMC allows for the prioritisation of somatic mutations that introduce biologically relevant changes to protein function, and participate in the development of cancer.
Metrics including ClinVar significance, dN/dS ratios, and variant frequencies in normal populations (gnomAD) have been integrated into this resource. They have been used alongside COSMIC data on mutations' prevalence across 1,500 forms of human cancer. This helps to predict candidates for driver mutations in the coding portion of the genome.
What does The Cancer Mutation Census allow me to do?
Searching for a gene returns a series of interactive graphs allowing you explore all COSMIC coding mutations in the context of the whole gene. Data are presented as per codon (AA residue).
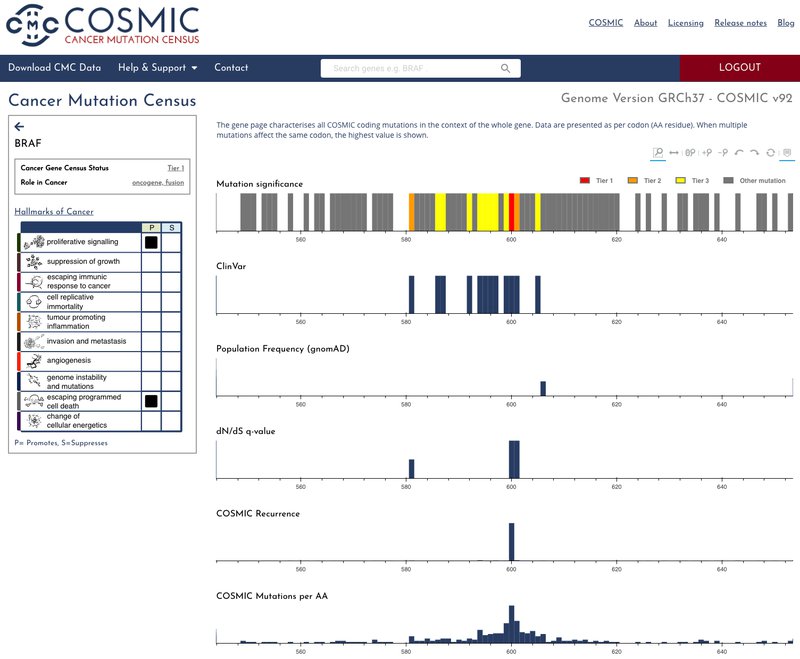
Clicking through on the amino acid position, brings you to the codon page where it is possible to scrutinise all COSMIC mutations that affect a selected codon. We have colour coded the codon graph backgrounds to denote their tier status, tier one (red), tier 2 (orange), tier 3 (yellow), other mutations (grey). By default the mutations are ranked in order of CMC significance but it is possible to filter and sort the results to query the data with your own requirements, for instance sorting by population frequency in descending order or filtering the codons to show those with a Clinvar evidence level above 4.
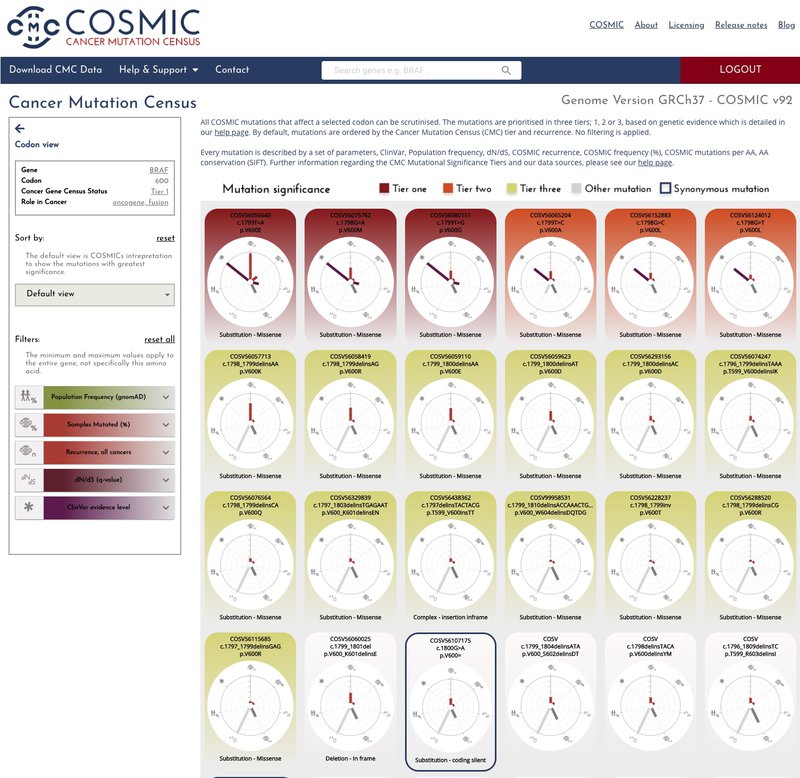
The mutation page allows you to delve deeper into the data by exploring the data that comprises the given CMC mutation significance tier.
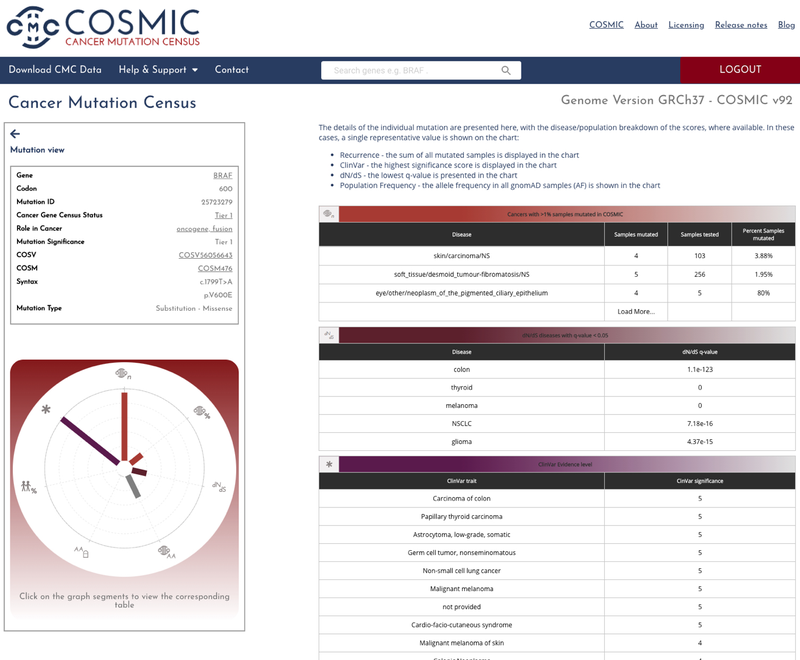
The CMC integrates data from the Cancer Gene Census, Hallmarks of Cancer and provides links back to the main COSMIC website allowing you to explore the vast amount of high quality manually curated genic and genomic data.
Currently the data is only available on GRCh37 for both the website and the download file, but we are working on GRCh38 and will make this available in a later release.
How do I access The Cancer Mutation Census?
The CMC is now live on https://cancer.sanger.ac.uk/cmc/, access it now!
To access the CMC you will need to log in using your normal COSMIC credentials. If you are new to COSMIC you will need to register at https://cancer.sanger.ac.uk/cosmic/register. If you require a license, you can find more information on our licensing pages, Licensing and Licensing FAQs.
Anything else?
For optimal viewing, please view the CMC on a screen no smaller than 1200px wide.
We’d love to hear your feedback! You may notice a few pop ups on the site as you explore the data, please take a moment to have your say so that we can continue to develop the CMC, building on the suite of data and tools COSMIC has to offer.
Access the CMC now!
Keep up date to with COSMIC
Please follow us on LinkedIn, Twitter, Facebook and via our email announcements to keep abreast of the latest COSMIC news and developments. If you do not receive our emails, please update your email preferences, as shown below, in your COSMIC account.

