From diagnosis to remission- Haematological cancer research to look out for in v97
22 Nov 2022
Haematological cancers arise from blood forming tissue such as cells of the immune system or bone marrow. Collectively, these are the fifth most common type of cancer in the world and account for 7% of all cancer deaths, with 1.24 million occurrences and 720,000 mortalities per year. Our COSMIC team have been expertly curating information on all groups in preparation for our v97 release.
So what differentiates the main haematological cancer groups?
Leukaemias occur in blood cells and bone marrow, they involve mutated leukocytes gradually outnumbering and suppressing development of normal ones.
Lymphomas develop in organs within the lymphatic system. Lymphoma is separated into two further groups, one of which (Hodgkin lymphoma) is one of the most curable forms of cancer.
Myelomas are a group of cancers that involve abnormal plasma cells and start in the bone marrow . Part of this abnormal behaviour involves the production of mutated antibodies.
Myeoloproliferative neoplasms (MPN) occur when the body produces too many of a particular blood cell, there are multiple types of MPN including polycythaemia, essential thrombocythaemia & myelofibrosis. Each type is defined by which blood cell is being overproduced.
Myelodysplastic syndrome is a disease characterised by insufficient healthy blood cells as a result of immature blood cells in bone marrow not maturing correctly.
Although these groups are broadly used for classification, it’s much more complex than that. As of COSMIC v97, we've recorded at least 340 unique forms of haematopoietic & lymphoid neoplasms and more than 6400 precise forms of human cancer, numbers that are growing constantly as ‘omics technologies become more advanced. Currently, WHO defines more than 100-clinically meaningful subtypes of haematological malignancies based on their presumed cell of origin, genetic abnormalities, and clinical features
From breast cancer to rare pancreatic cancers, COSMIC updates have spotlighted a range of diseases. The decision to focus on a particular group of cancers is never taken lightly, with countless countless considerations to be made. How much novel data is available? Have new sub-classifications been published? What other groups have we focused on recently?
After careful contemplation of these factors and more, haematological cancers were chosen as v97’s focus.
So what did the team find? Here’s a sneak peak into three papers curated as part of the v97 update, each putting the spotlight on distinctive areas of research improving the outcomes for haematological cancer patients.
A novel mutation & how to tackle it
When genes are known to be recurrently mutated and drivers of cancer, they become high priority for targeted therapy research. But what happens when a novel activating mutation is discovered in one of these genes? Will previously discovered therapies still be of clinical use?
Bruno et al 2021 found precisely this in fms-related tyrosine kinase 3 (FLT3), one of the most common driver genes for acute myeloid leukaemia (AML). FLT3 acts as a class III receptor tyrosine kinase and is involved in the regulation of cell death (apoptosis), cell division (proliferation), and differentiation of hematopoietic cells. Mutations in FLT3 are responsible for almost a third of all acute myeloid leukaemia cases, and this gene is used in the molecular genetic diagnosis and risk stratification of AML. Hence, a targeted therapy is already available for newly diagnosed patients - the tyrosine kinase inhibitor, midostaurin.
In their case report, Bruno et al, describe a 32 year old woman who presented with AML. Next Generation Sequencing (NGS) analysis revealed an in-frame deletion in FLT3, and after consulting COSMIC , it became apparent that this had not been reported before. This particular in-frame deletion (c.1770_1784del15; p.Phe590_Arg595delinsLeu) affects Exon 14, which codes for the autoinhibitory juxtamembrane domain. The novel FLT3 variant leads to constant activation of the FLT3 receptor - with downstream activation of pathways that could aid the cells to grow and evade death.
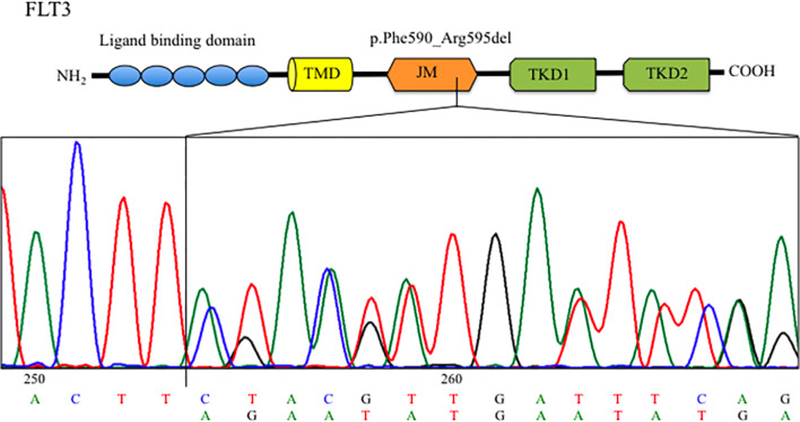
The patient in question responded well to conventional chemotherapy and transplant. But what if she hadn’t? And what about other patients who may present with this mutation in future? Rather than leaving their research at discovery, the researchers went on to test whether the mutation was sensitive to midostaurin. They succeeded in demonstrating that the drug reduced the clonogenic capacity in primary blast cells, offering useful information for clinicians treating AML patients and illustrating the need to characterise genomic alterations of all AML patients.
Classification of contradictions: A new system for TET2
Since haematological cancer covers such a vast range of diseases, there is an equally vast range of mutations with pathogenic consequences. Identifying these mutations, their implications, and actionability can be crucial to a patient’s wellbeing, so how do we begin to make sense of all of this data?
TET2 encodes an enzyme that catalyses the conversion of methylcytosine to 5-hydroxymethylcytosine - or in other words it helps transfer an oxygen atom to the methyl group of 5-methylcytocine. The TET family of enzymes play an important role in epigenetic modification and play an important role in transcription regulation, but TET2 itself is involved in many more cellular processes including resolution of inflammation, mast cell differentiation, and neurogenic rejuvenation. TET2 is expressed at particularly high levels in hematopoietic cells, playing a vital role in hematopoiesis. Mutations have been reported in haematological and lymphoid neoplasms, myeloid neoplasia, and age-related clonal hematopoiesis as well as others. Yet previous attempts to use the TET2 gene as a predictor of clinical variables have led to contradictory results. The overwhelming number of mutations reported make it difficult to interpret new variants or understand the functional consequences.
Bussaglia et al 2019 tackled this issue head-on. They analysed the many missense mutations within TET2 that are linked to uncontrolled and abnormal cell growth (neoplasia). For this study, samples were gathered from 189 AML patients and analysed following Sanger and NGS. Next, it was scrutinised bioinformatically alongside existing data for the TET2 gene, some from COSMIC.
Thirty-eight TET2 allelic variants were identified as part of the study, four of which were reported for the first time:
- COSM10132810 c.122C>T (p.P41L)
- COSM10132814 c.4535C>G (p.A1512G)
- COSM10132815 c.4760A>G (p.D1587G)
- COSM10132816 c.5087A>T (p.Y1696F)
Using this data, the researchers developed a classification system, taking into account characteristics of the mutations such as possible structural implications and sequence conservation.
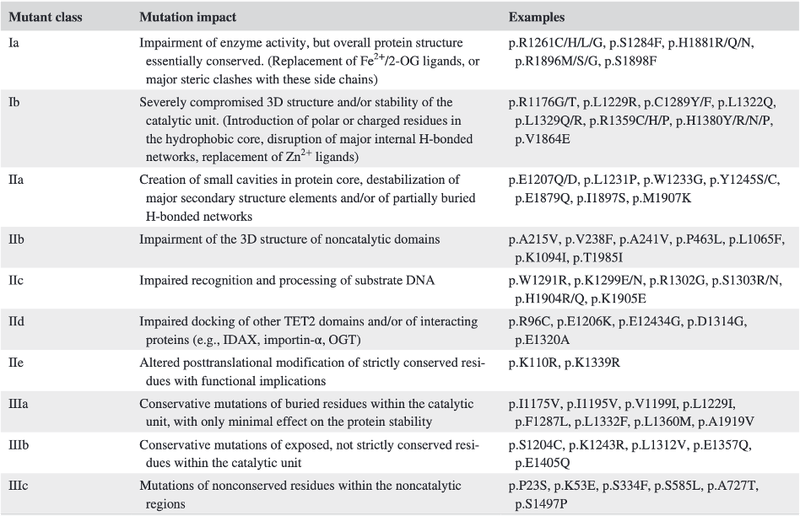
After years of contradictory research into the consequences of TET2 mutations as a whole, Bussaglia et al ’s proposed classification system allows each mutation to be considered individually. This ability to quickly define a mutation by in vivo consequences has the potential to assist in the design of advanced immunotherapeutic approaches and effect prediction of prognosis for leukaemia patients in the future.
Prediction of treatment response and long term health
Cancer is often discussed colloquially as if it is one disease, but in COSMIC alone there is data on over 6000 unique cancers, and this is what makes targeted drug design invaluable to patient outcomes. It is widely acknowledged that these targeted treatments can often help patients with a wide range of pathogenic mutations, but to varying levels of success.
And of course, other factors come into play. In particular, age. Older patients (65 or older) with acute myeloid leukaemia (AML) and who don’t qualify for standard treatment have an extremely poor prognosis - fewer than 10% survive beyond 3 years. Likely because of unfavourable genetic profiles of elderly patients, comorbidities that aggravate therapy side-effects and high relapse rates. But this has been shifting in recent years with the introduction of new targeted therapies, such as the FLT3 inhibitors mentioned earlier. But to maximise treatment efficacy, it is imperative to develop biomarkers to guide treatment approaches in elderly patients.
Which raises the question: Could we predict a patient’s response to treatment based on the pathogenic mutation they are carrying?
Ayala et al carried out mutational profiling on bone marrow samples from a cohort of elderly AML patients taking part in the FLUGAZA phase III clinical trial (PETHEMA group). They used a customised 43-gene NGS panel for myeloid pathology, analysed the results, and patients were treated with azacytidine or fludarabine plus low-dose cytarabine.
The team found that the mutational profile of older patients with AML is different to that of younger patients, and several prognostic markers in young people have no impact in older patients. The outcomes of these treatments allowed the team to define a molecular signature for identifying responses to azacytidine treatment. An example of this includes certain EZH2 and IDH1/2 mutations being associated with a patient responding to the CpG methylation caused by azacytidine. Additionally they were able to identify, as part of the fludarabine plus low-dose cytarabine trial, that mutations in the BCOR gene was associated with a lower rate of relapse free survival. Research into this area could, in the future, allow for more accurate and faster selection of treatment for a disease where time is absolutely critical to survival.
From diagnosis to remission
With haematological cancers covering such a wide variety of unique diseases, seeing the breadth of research being carried out is encouraging. Studies such as the ones above have the potential to offer those working with patients one of the most crucial resources available: Time. Being able to quickly identify the best course of action for treatment of an individual’s cancer is imperative to patient outcomes and often the difference between life and death.
The research that offers caregivers this time would not be possible without a trusted and vast source of data, curated by experts with a vested passion.
This is why we’re proud to launch v97 in November 2022, a COSMIC update with a focus on haematological cancer data. For further updates on the release and all things COSMIC, you can opt in to marketing communications via your account or follow us on twitter and linkedin.
