
COSMIC Release v91 is live!
7 Apr 2020
Head over to our release notes to find a detailed description of the content of this release, including a summary written by our expert curators for the genes in focus for this release.
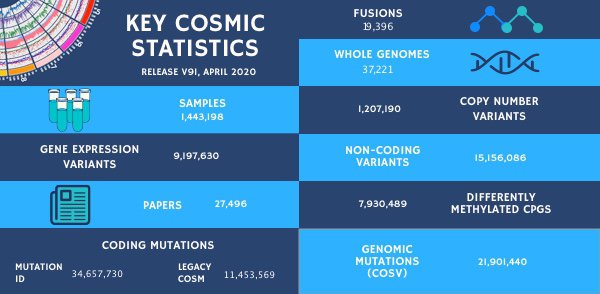
Summary of our updates for v91
- We have performed high-quality manual curation on 4 new cancer genes which are TRRAP, KMT2A, MYC and CDKN1B. There are substantial updates to the existing expert curated gene APC. We have also focussed on testicular and other male cancers as well as breast implant related lymphoma.
- We have also updated our ICGC dataset to v28, which includes 2 new studies: ICGC (BPLL-FR): B-Cell Prolymphocytic Leukemia - FR and ICGC (GACA-JP): Gastric Cancer - JP; with a complete re-annotation using Ensembl Variant Effect Predictor (VEP).
- In response to user feedback, we have provided a new mutation tracking file for COSMIC and the Cell Line Project to map the legacy COSM/COSN IDs to the new genomic ID (COSV), along with the gene names, accession number and a new unique mutation identifier. We also indicate in the file if these mutations are coding or non-coding. There is also a field that indicates if the annotation is on the canonical transcript.
- We have improved our VCF files to include HGVS syntaxes on the genomic (HGVSG), on the CDS (HGVSC) to include the transcript accession number with the version, and on the peptide (HGVSP) with Ensembl's peptide accession.
- To our VCF files, we have added a normalised version denoted with the suffix <normal>, where each variant is 5' shifted whilst maintaining the HGVS compliant (3’ shifted) syntaxes in the INFO section. This reflects the non-normalised version where different. We have also compressed these files with bgzip following user feedback.
- All our files with the mutation syntaxes have got these additional columns of HGVSG, HGVSC and HGVSP.
If you have any suggestions for content or functionality that you would like to see in a coming release, please contact us at cosmic@sanger.ac.uk.
