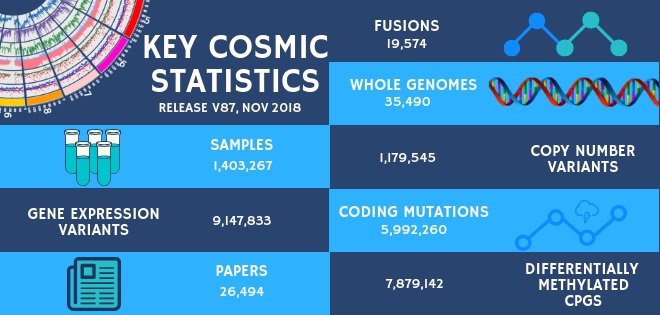
COSMIC Release v87
13 Nov 2018
The November COSMIC release (v87) is now live! We have 4 newly expert-curated genes: ARID1B, RBM10, B2M and BCORL1; a significant update to KRAS; and the newly curated fusion pair ETV6-PDGFRB. We have continued the disease focussed curation trialled in the last release and this time we have updated the expert-curated mutation data for mesothelioma, a rare and aggressive cancer.
We've incorporated samples from 9 systematic screen papers. There have been 4 new genes added to the CGC and also 8 new genes that have Hallmarks of Cancer. Several new samples and drug/gene combinations have been added in the drug resistance section. Finally, we've been writing papers on the new developments happening within COSMIC, so make sure you have a read.
New genes: ARID1B - AT-rich interaction domain 1B, like ARID1A, encodes a protein which is a component of the SWI/SNF chromatin remodelling complex and may play a role in cell-cycle activation. Somatic mutations in both genes are very frequent in gynaecological and several other solid tumors. ARID1B mutations are found in approximately 20% of ovarian clear cell carcinomas and are also detected in dedifferentiated ovarian and endometrial carcinomas where concurrent ARID1A and ARID1B inactivating mutations result in loss of protein expression in 25% of the tumours. ARID1B mutations have also been found in various other tumours, as detailed in full here. ARID1B may be targetable with FDA-approved HDAC inhibitors, including vorinostat and panobinostat.
RBM10 - RNA binding motif protein 10, encodes a spliceosomal protein involved in the regulation of gene expression. Located on the X chromosome (Xp11.3), mutations have been found in a variety of cancer types, including breast, colon, thyroid, ovary, pancreas, prostate and lung. The genetic changes are mainly missense SNVs, although frameshift insertions predicted to generate truncated proteins, and nonsense mutations, have also been found. RBM10 acts as a tumour suppressor gene and these loss-of-function mutations affect the mechanism of repression of Notch signalling and cell proliferation through the regulation of NUMB alternative splicing. Mutations in RBM10 have also been implicated in the drug resistance mechanism of thyroid carcinoma with the BRAF V600E mutation.
B2M - beta-2-microglobulin, encodes for the constant light chain of the classical major histocompatibility complex (MHC) class I molecule. Somatic mutations in B2M, which include substitutions, deletions and LOH of chromosome 15q21, inhibit transcription of B2M or affect translation of the mRNA, resulting in a lowered expression of the protein or in synthesis of a non-functional protein. Mutations are spread across B2M, however, a frequent mutation at the position M1 seems to be recurrent in lymphomas and a two nucleotide deletion in a CT-repeat in colorectal carcinomas. Decreased expression of B2M has been reported to be associated with worse prognosis in non-Hodgkin lymphoma patients and a favourable prognosis in Hodgkin lymphoma patients. Truncating B2M mutations may be associated with acquired resistance to PD-1 blockade in metastatic melanoma.
BCORL1 - BCL6 corepressor like 1, Xq25-26.1, is homologous to the tumour suppressor gene BCOR and ubiquitously expressed in human tissues. Somatic mutations have been found in a variety of myeloid tumours including acute myeloid leukaemia, myelodysplastic syndrome and chronic myelomonocytic leukaemia. Frameshift mutations are most common, although missense mutations also occur. Splice site or nonsense mutations across the gene are predicted to result in severely shortened truncated proteins lacking the LXXLL nuclear receptor recruitment motif and the C-terminus. Similar somatic mutations have also been reported in solid tumours such as MSI-H gastric adenocarcinoma, melanoma, Wilms tumour, intracranial germ cell tumours, gliomas and head and neck squamous cell carcinoma.
Update to KRAS - Kirsten rat sarcoma viral oncogene homolog, was one of the first 4 genes that was curated for COSMIC when it was released nearly 15 years ago. Over the last decade, KRAS has become one of the most clinically important and sequenced oncogenes in cancer. Accordingly, the related scientific literature in PubMed has exploded to a level that is impossible to manually curate exhaustively. With the help of PubTator and LitVar powered by machine learning (Wei, 2013; Allot 2018), we have scanned the literature from the last 5 years and managed to curate 21 new mutations for this COSMIC release. These consisted of 15 new substitution mutations, 1 nonsense mutation and 5 new deletions/insertions from 17 publications. In total, 70 new KRAS mutations have been added to COSMIC during 2018. Most of the mutations are found outside the well-known oncogenic hotspots expanding the number of potentially relevant mutations in oncology.
ETV6-PDGFRB - the rare fusion ETV6 (TEL)-PDGFRB, the molecular consequence of the t(5;12)(q33;p13) translocation, is found in some patients with chronic myelomonocytic leukaemia and other myeloproliferative disorders with eosinophilia. ETV6 encodes an ETS family transcription factor containing a Helix-Loop-Helix (HLH) and an ETS DNA binding domain. PDGFRB encodes a cell surface tyrosine kinase receptor for members of the platelet-derived growth factor family. In the fusion, the N-terminal HLH domain of ETV6 is fused to the transmembrane and the tyrosine kinase domains of PDGFRB. The breakpoints detected in ETV6-PDGFRB transcripts are consistently at exon 4 in ETV6 and at exon 11 in PDGFRB. Imatinib is effective therapy in patients with ETV6-PDGFRB-positive chronic myeloproliferative diseases.
In addition to the curated genes, 4 new genes added to the Cancer Gene Census, LATS1 and LATS2 into Tier 1, and MACC1 and SETDB1 to tier 2. Additionally, 8 new genes added to the Hallmarks of Cancer. These are CYLD, LATS1, LATS2, MEN1, MYD88, NOTCH1, NOTCH2, and RUNX1T1. There are 9 new systematic screens in version 87, for further technical details please see the release notes.
