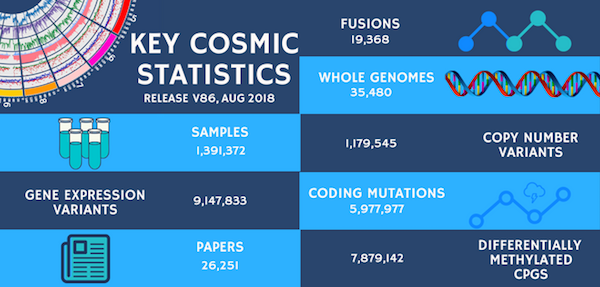
COSMIC Release v86
14 Aug 2018
The August COSMIC release (v86) has just gone live! We have 3 newly curated genes: CHD4, IRS4 and CTCF; and the newly curated fusion pair MN1-ETV6. As part of this release we have also completed a body of curation focused on Glioblastoma
We've incorporated 476 whole genome screened samples from 8 systematic screen papers along with 1550 from ICGC Release 27.There are also 7 new genes added to the Hallmarks of Cancer.
CHD4 - CHD4 (chromodomain helicase DNA-binding protein 4) encodes a protein belonging to the SNF2/RAD54 helicase family and which is the main component of the nucleosome remodelling and deacetylase complex, with an important role in epigenetic transcriptional repression. A high frequency of mutations in CHD4 (17%) has been found in serous endometrial carcinoma where most are missense mutations and half affect the ATPase/helicase and helicase domains. CHD4 is also mutated in clear cell, endometrioid and mixed-histology endometrial tumours, and in uterine carcinosarcoma.
IRS4 - IRS4 (insulin receptor substrate 4) is a cytoplasmic scaffold protein that is phosphorylated by the insulin receptor tyrosine kinase in response to receptor stimulation by insulin. Tyrosine phosphorylated IRS4 protein has been shown to associate with cytoplasmic signalling molecules that contain SH2 domains leading to the activation of the P13K/AKT and MAPK/ERK signalling pathways. The gene for IRS4 is located on chromosome Xq22.3 and somatic mutations, including point mutations and deletions, have been found in a variety of tumour types including metastatic melanoma, multiple meningioma, paediatric T-cell acute lymphoblastic leukaemia and other haematological malignancies. An insertional mutagenesis study in mouse has shown that IRS4 is a driver in mammary oncogenesis, working synergistically with ERBB2/HER2. Furthermore, analysis of expression of IRS4 in human breast carcinomas has shown it to be a putative biomarker for HER2-targeted therapy resistance.
CTCF - CTCF (CCCTC-binding factor) encodes a transcriptional regulator affecting chromatin structure and organisation by binding different DNA target sequences and proteins, thus playing a vital role in transcription by controlling promoter-enhancer interactions. Mutations have been observed in a variety of cancers including endometrial carcinoma (and the potentially precancerous lesion endometriosis), bladder cancer, Wilms tumour, several myeloid tumours (including acute megakaryoblastic leukaemia (AMKL), transient abnormal myelopoiesis preceding AMKL in Down syndrome patients, and acute lymphoblastic leukaemia), head and neck cancers and some breast cancers. CTCF functions as a tumour suppressor in cancer. Most mutations are frameshift, nonsense or splice mutations resulting in haploinsufficiency following nonsense mediated decay, or are loss-of-function missense mutations often in the zinc finger domains. However, some missense mutations alter DNA binding residues and act as gain-of-function mutations enhancing cell survival, for example p.K365T in endometrial carcinoma.
MN1-ETV6 - The MN1-ETV6 fusion, resulting from t(12;22)(p13;q12), is a recurrent but infrequent abnormality in haematological malignancies. MN1 encodes a transcription co-factor while ETV6 encodes a protein of the ETS transcription factor family. Two fusion transcript types have been reported and in both most of MN1, including the glutamine/proline rich domain, is fused to the DNA binding domain of ETV6. In type I, exon 1 of MN1 is fused to exon 3 of ETV6 and in type II the same MN1 exon is fused to ETV6 exon 4. MN1-ETV6 fusions are found in myeloid leukaemia and myelodysplastic syndromes.
There are also 7 new genes added to the Hallmarks of Cancer.DICER1, IDH1, IDH2, KIT, NF1, NF2 and PIK3R1.
There are 8 new systematic screens in version 86 COSP33196, COSP43017, COSP43572, COSP43892, COSP44274, COSP44557, COSP44783 and COSP44991.
COSMIC has now integrated ICGC release 27 from April 30th 2018 adding 1 new study COSU657 WT-US and updating 4 COSU340 CLLE-ES, COSU382 PACA-CA, COSU537 PRAD-CA and COSU538 PRAD-UK.
