
COSMIC Actionability: Release v93 (March 2021)!
24 Mar 2021

We are proud to announce after three years of high quality manual curation from PhD level experts, our latest product in the COSMIC suite, Mutation Actionability in Precision Oncology (Actionability), has launched.
At COSMIC we pride ourselves on 50+ person years of high quality manually curated data by PhD level experts which allows researchers to understand the genetics that drive cancer, which are the genes and mutations that matter and finally, seek to understand what can be done about them.
The Actionability journey started in 2018 when we sought to provide a resource that addresses the question “What treatment options are available or under investigation for cancer patients with somatic mutations?”, adding another piece to the puzzle of precision oncology.
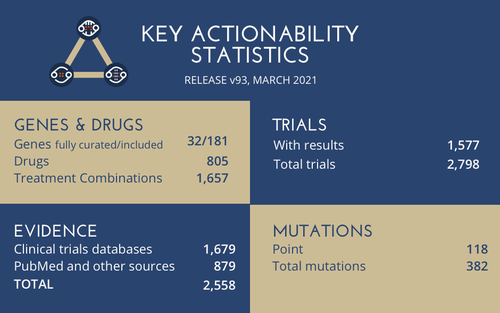
The aim of COSMIC Actionability is to indicate the availability of drugs that target mutations in cancer and track the progress of clinical studies towards making new drugs available. Drugs that target somatic mutations are represented at all stages of drug development, through safety and clinical phases to market and repurposing, with additional case studies. Primarily the data comes from clinical trials databases, however our manual curation efforts add up to 3x more data than using clinical trials alone*.
The principal units of actionability are mutation, disease, and drug. Capturing relations between these units allows identification of existing and upcoming drugs that target particular genetic variants in specific cancer types.
In our first release of Actionability (v93), the data includes well-known genes such as EGFR and KRAS but also other genes such as EZH2 for its topical scientific relevance. We have curated point mutations, fusions and other mutations.
A full list of gene coverage can be found in the Actionability about page.
Our release cycle will follow that of COSMIC, three times per year. At every release, we aim to update the existing records to ensure that the results are up to date whilst adding curating additional data.
We have been thrilled at the responses to our request for feedback on the pre-release sample data file and the interactive prototype of the website. We are responding to your feedback. Future versions of Actionability will extend mutation mapping and have closer integration with other COSMIC tools. We’d love to hear your feedback and if you have any thoughts for the future direction of Actionability, please contact us via cosmicengagement@sanger.ac.uk.
The Actionability data is available as a download file containing the entire actionability dataset, from the COSMIC downloads page. More information can be found on the Actionability about and help documentation pages.
For academic use, the data is freely available. For commercial use of the data, a license fee may apply, to find out more please contact our commercial distribution partner, Qiagen via bioinformaticssales@qiagen.com.
*Calculation can be found in FAQs.
Keep up date to with COSMIC
Please follow us on LinkedIn, Twitter, Facebook and via our email announcements to keep abreast of the latest COSMIC news and developments. If you do not receive our emails, please update your email preferences, as shown below, in your COSMIC account.

