Copy Number Signatures: A scalable research and clinical tool?
15 Jun 2022
Copy number signatures (CN Signatures) are the latest addition to COSMIC Mutational Signatures, a reference database of characteristic combinations of mutation types arising from specific mutagenesis processes. Ahead of the paper release, I caught up with one of the leads, Dr Ludmil Alexandrov, to discuss the key findings, utility of the data, and hopes for the future.
Ludmil is a Wellcome Sanger Institute alumnus and is now based at UC San Diego as an Associate Professor in Cellular and Molecular Medicine. His strong computational background lends itself to his research interests of leveraging the information hidden in large-scale omics data for better understanding of the mutational processes causing human cancer, for identifying potential cancer prevention strategies, and for developing novel approaches for targeted cancer treatment.

Q1. Can you give me a one sentence summary of the paper: Signatures of copy number alterations in human cancer?
We have known for a long time that copy number changes are important in cancer. And in this paper, we provide the first comprehensive blueprint of the common patterns of copy number changes across cancer.
Q2. Well that leads quite nicely into my next question: Why are copy number changes important in cancer?
Well, copy number changes embody any parts of the chromosomes being either lost or gained. If you lose a tumour suppressor gene, then it stops suppressing the tumour development, and that can lead to cancer. And if you gain a gene that has proliferative abilities, then it's going to make the cancer proliferate.
We’ve known that these changes have been very, very important for a long time. But we haven't really understood the processes that generate them. Essentially, this study digs into that specific topic, it says, ‘let's look at what patterns we have and the genomic scars left by this copy number change, then use that to figure out the processes that are generating these specific changes’.
Q3. Can you explain some of the processes attributable to the copy number variants signatures?
Sure, let me go through them one at a time.
Ploidy
Normally, you’d expect two copies of a chromosome in a normal somatic human cell – one from the mother and one from the father. However, in cancer cells, there may be four copies of each chromosome or eight copies or a very large number. So this is described as the ploidy, the amplification of the whole genome and the process that caused it.
Amplification
This refers to specific parts of the chromosome being amplified, or increased, which can result in hundreds of copies of this one small region. If it's in a region containing an oncogene, that can lead to proliferation.
Loss of heterozygosity
The first time I was learning about these, I was like, ‘what on earth is that?!‘ My understanding since then has evolved! So typically you have two copies of each chromosome. Generally, loss of heterozygosity is when you lose one of the copies of that chromosome. So now you're left with just one. It can be more complicated but that’s the basic description.
Chromothripsis
Chromothripsis occurs when a chromosome just shatters into many tiny pieces. And then in one way or another, it gets assembled but in a jumbled way. It’s a shattering and then assembling in a somewhat random manner.
Tandem duplications
The way I always imagine these is like jumping frogs. It’s a segment of the DNA that just jumps and jumps and jumps, and duplicates and duplicates and duplicates, and they happen to do it together. So you end up with long duplicating regions.
Q4. Okay, so I see how these copy number variants are occurring, but why is it important to consider them in the context of signatures?
Let’s start with the general importance of mutational signatures, we have found they can inform in three ways. First, they can inform the biology and explain what caused the specific pattern in a cancer. Secondly, they can be utilised for prevention. If you understand what causes the signature, and that cause is external, you can potentially prevent that exposure. Or you can monitor people who have germline polymorphisms that increase risk. Thirdly is clinical biomarkers, signatures can be used as predictive or prognostic biomarkers.
We already had that information for signatures in substitutions, indels, and doublets. But not for copy number changes - we were missing one of the major types of changes in the genome.
Now we have it. Now we can utilise copy number signatures for understanding the processes that cause them, for potentially preventing exposures, and as clinical biomarkers.
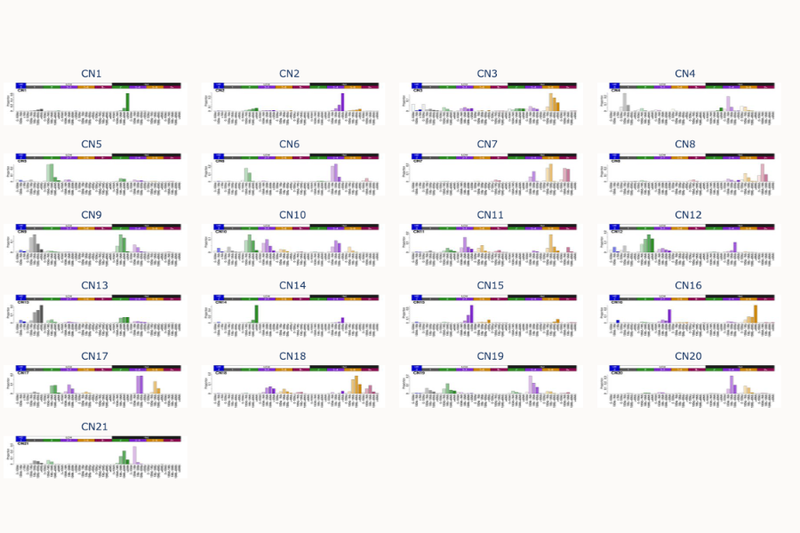
Q5. How many new signatures have you identified for copy number changes? And how did you identify them?
We have created a completely new reference set of signatures. We had thousands and thousands of genomes, and we have summarized and can explain them with 21 signatures.
How did we do it? Well, from a very high-level perspective, we use AI algorithms. And when one says AI, that's a lot of hand waving. So without going into too much detail, the AI algorithm we use is very similar to the one Netflix uses to give recommendations for movies. Our process tries to summarise a lot of information into meaningful subsets and then find the ones that may be most useful.
Q6. And what data did you feed it to come out with those recommendations?
For this paper, the reference set of signatures is based on data from the Cancer Genome Atlas, the TCGA.
Q7. So in theory, people can get their own data from their own studies. They can feed it in and the ‘math methodology/algorithms’ allows them to identify signatures in their own data?
Absolutely. And it’s important to keep in mind that copy number signatures are unique because they can be applied on whole genome or whole exome sequencing, SNP-6 microarrays, single cell sequencing, and reduced representation sequencing. So they can be applied with virtually the same resolution across these different profiling platforms.
Now, in contrast, previous signatures - such as single base substitution signatures - have much higher resolution when applied to genomes than when they are applied to exomes. With copy number signatures we have very similar resolutions, which means they can be applied easily across many fields. For example, in clinical fields where they use panels and exomes, or in research fields where they use genomes, or even fields such as very large scale epidemiology where they do microarrays.
The other cool thing about this paper is we're not only doing the blueprint and the references that are seen in COSMIC, we're also providing the tools. You can have the data, you can plug it in, and compare it with COSMIC Signatures. So there’s functionality allowing researchers to immediately use these CN signatures
Q8. Moving onto some of the detail in the paper, you discuss macro-level events like genome doubling and how this leads to evolution in the type of copy number signatures. Can you explain that?
This is again where CN signatures are slightly different to previous signatures. With single base substitutions, there are three-billion possible locations where mutations can fall around the genome, or six-billion when you consider two copies. So the chances of them overlapping with one other are very minor.
When you have a copy number changes affecting the whole genome or a whole chromosome, the chances of these things overlapping becomes much higher. In fact, this is fairly common. We see that if you have a particular signature, and then the genome doubles, the signature evolves into a different one. This evolution continues as the doubling continues.
Q9. Is this a one directional unpredictable thing? So is it something that could be predictive of disease progression?
Not necessarily as there are multiple different paths. The first path may be a double-double, and then some losses and so on. There are multiple ways to get to the same place, always with some interaction. The final result we see is the sum of those events.
So we don't know whether they’re clinically meaningful and we don’t have serial samples (yet).
But they do make quite an interesting pattern on the graphs, even if I didn't necessarily understand them…
(Laughs) I think genomics people are guilty of doing graphs that are intimidating to others, just so they're completely overwhelming. Nobody understands and you just move past it to say, ‘Okay, this must be correct…’
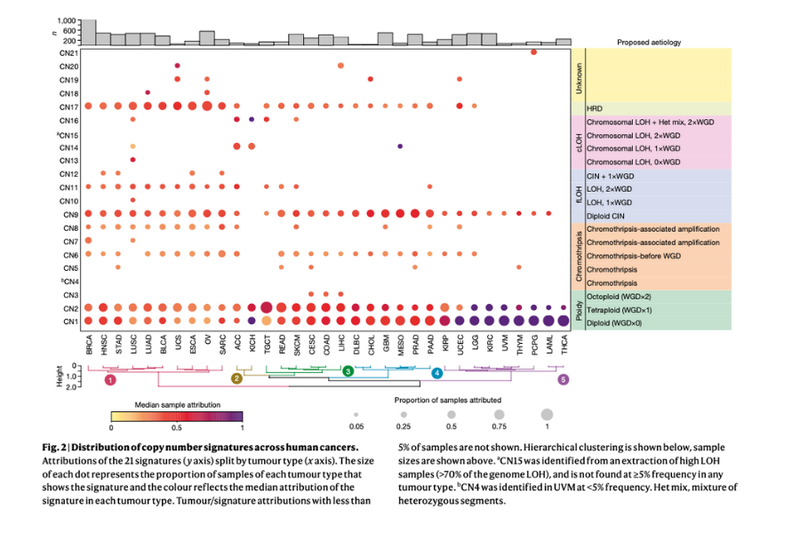
Q10. So you also found that there were certain signatures that were specific to certain cancers? What are the key takeaways from that? Maybe you could talk through a particularly interesting example?
There were some very clear copy number signatures across many cancer types, which tells us about the common events. But as you said, there were signatures that are confined to a set of cancer types and even a small subset of cancer types. Some of them unfortunately, we don't understand. We have one example within a small subset of uveal melanomas. It's very clear it's there, but we just don't know what causes it.
The example we do understand is the signature of homologous recombination deficiency and tandem duplications, which is in a subset of cancers including the BRCA deficient breast and ovarian cancers. We understand why it's there - the repair pathway has failed in these cancers. This is getting generated and can be used as a readout for clinical biomarkers.
Q11. Is there any benefit to analysing copy number signatures alongside other types of signatures? Are they complementary?
There’s definitely some that are complementary. For example, I mentioned the homologous recombination deficiency, we have indel and SBS signatures that are very highly correlated with one another.
But there's also exceptions, places where we only see the copy number signatures. And that probably says something about interesting biology, and vice versa.
There’s also the point of utility, because other signatures require whole genomes. And whole genomes are not typically used in places like clinical settings. Now, the copy number signatures can be applied to most of the clinically available data. So that gives you this complimentary type of analysis that can be utilised very fast within a clinical setting. And we have already shown a number of prognostic biomarkers.
Q12. There seems to be a growing theme to research efforts around health equity. And the fact that, realistically, if we're going to have these kinds of solutions clinically, it needs to be a lower resolution approach that doesn’t require the whole exome or whole genome approach? Does this fit into that?
Exactly, in the US, there are some ASCO reports about the standard-of-care biomarkers for treatment, like KRAS mutations in lung cancer. These were discovered 2004-2005, and introduced as the standard of care. Yet in the US, less than 50% of people get screened for them. Globally, less than 5% of people get screened.
So we have something that we know will save lives. Yet even in one of the richest countries less than half get screened and worldwide essentially no one gets screened.
It's a bit sad, because yes - it's great science. And we say yes - it's translated. But it's actually translated for a very small proportion of people who can afford it. That’s why work like this is so very important.
Q13. And that segues nicely into how these results and signatures fit into the bigger picture of tackling cancer?
From a wider perspective, the basic science is very clear. The DNA repair expert can use the CN Signatures to disentangle and answer questions like: What repair processes are happening? What's causing these different events? What is the damage? Why does the cell get these signatures? How does it survive? So it just gives another apparatus to tackle this problem, and I think it will be quite a powerful one.
But I'm particularly excited about the clinical utility, because we know that they're prognostic. The example is the case with many amplicons. In many cases, that's indicative of things like extra chromosomal DNA - this circular DNA that comes out of the chromosomes and is amplified. The signature captures that and can be utilised to demonstrate this event in clinical settings.
We have a number of these signatures that can immediately get fed into existing clinical pipelines to give better predictions of patient survival. Although we haven't yet shown anything that predicts response for specific drugs.
Q14. Are there particular signatures that you think are more likely to be used in that context?
There are two examples that are very clear. Firstly are the homologous recombination deficiency signatures. We're developing this into something that can be utilised in a much more inclusive manner - including if you don’t have whole genomes. This signature may be useful in regards to predicting response to PARP inhibition.
Secondly is the amplicon signature that I previously mentioned. Several groups are now developing drugs to specifically target this extra-chromosomal DNA - but how will they detect it’s there in the first place? Well we provide that first step. Our amplicon signature could be used to predict the presence of extra-chromosomal DNA and impact on prognosis.
Q15. Does it change how actionable things are when they're driven by endogenous versus exogenous events?
Yes, because exogenous events are very useful for epidemiology and for screening large populations. Endogenous ones tend to be more useful for therapeutics, because if something has happened in the cell, and is potentially ongoing, which means that there is vulnerability in that cell. So endogenous ones may be more useful for targeted specific treatments, whereas the exogenous ones can be used for cancer prevention.
Q16. And a quick final question - what's next for you?
We hope to keep expanding our existing signature sets and pushing the tools that analyse them.
But actually we're now moving heavily into engineering them into useful and scalable solutions. We know they’re valuable. But we also know that to apply them to five or six-thousand people, one needs to spend around 20 million. The question is, can we make them useful if you spend something like 10 dollars per patient? So we're putting a big emphasis on that at the moment and moving into the translational space to make them useful and applicable to everybody.
You can read the paper, Signatures of copy number alterations in human cancer, here.
You can access the copy number signatures here.
- By Rebecca White, Head of Scientific Communications for COSMIC
