Bile Duct Cancer (Cholangiocarcinoma) Focus
15 May 2019
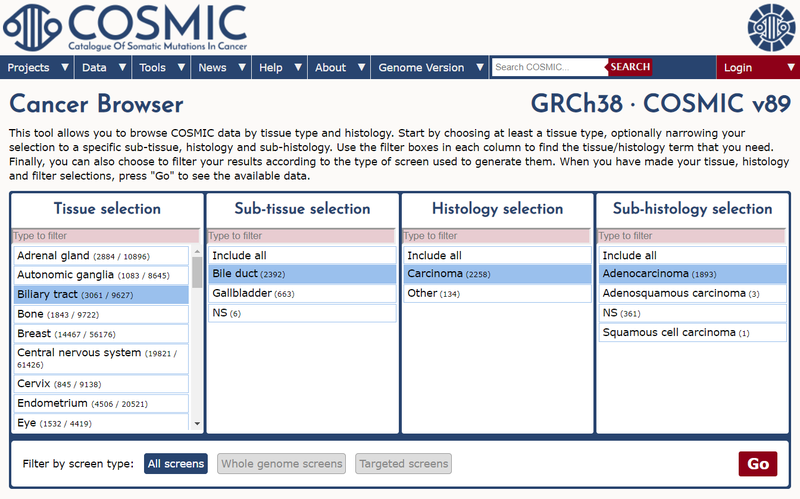
As part of release v89 we have focused on updating the expert-curated mutation data for bile duct cancer (cholangiocarcinoma). Approximately 60 additional publications that include mutation screening data in this disease are included in the release.
Bile duct cancer (BDC) is the second most common adult primary liver cancer after hepatocellular carcinoma and the majority are adenocarcinomas arising from the epithelial lining of the bile duct. They are classified as intrahepatic, perihilar or distal extrahepatic based on the predominant location, and each subtype has a distinct epidemiology, biology, prognosis and disease management protocol. There is a range of associated risk factors for BDC including primary sclerosing cholangitis, hepatitis B and C, and in South East Asia, particularly northern Thailand, liver fluke infection. Worldwide the incidence of intrahepatic BDC (iBDC) is increasing and its prognosis is dismal; patients with metastatic disease have a life expectancy of less than one year. Since iBDC remains asymptomatic until reaching an advanced stage most patients present with advanced, unresectable or metastatic disease, limiting the number of available treatment options. Surgical resection remains the mainstay of potentially curative treatment for localised BDC but even after surgery the prognosis is poor. Many patients are not suitable for resection and these cases may receive palliative treatment with gemcitabine plus cisplatin.
The genomic landscape of BDC differs depending on disease location. IDH1 and IDH2 mutations, mostly at their hotspots, are almost exclusive to iBDC, while ERBB2 and BAP1 mutations are common in extrahepatic BDC (eBDC) and less common in iBDC. Recently, FGFR2 alterations, mostly fusions, have been identified in iBDC. This update includes a paper by Churi et al. (COSP38231) who perform next generation sequencing on both iBDC and eBDC, demonstrating their significant molecular differences. In iBDC, genetic alterations in KRAS, TP53 or MAPK/mTOR are significantly associated with a worse prognosis while FGFR alterations correlate with a relatively indolent disease course. In a publication by Trachu et al. (COSP44522) molecular alterations are also explored, as well as clinical prognostic factors, in Thai patients with BDC. Goeppert et al. (COSP46197) use integrative genomic and epigenomic analysis to identify four major iBDC subgroups with genomic and epigenomic differences and prognostic implications.
Genomic studies of BDC have shown that they have many potential actionable targets. Zou et al. (COSP38150) identify 8 potential targetable driver genes, including IDH1, TP53, KRAS and ARID1A, in a Chinese population. In cases with FGFR2 fusions, promising therapeutic activity has been demonstrated with tyrosine kinase inhibitors such as BGJ398 that inhibit the FGFR2 pathway but such success is tempered by follow-up reports of resistance. Goyal et al. (COSP42875) describe genetic mechanisms of clinical acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive iBDC.
Publications in this update also cover the molecular characterisation of rare variants of BDC, including clear cell intrahepatic cholangiocarcinoma (COSP45941) and mucinous cholangiocarcinoma (COSP45954), as well as the combined tumour hepatocellular-cholangiocarcinoma (COSP42266).
